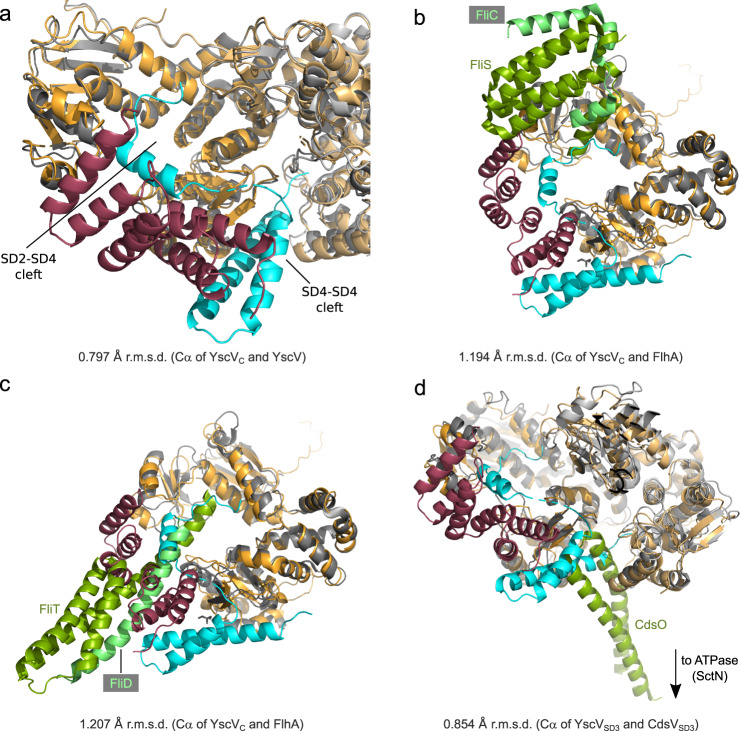
Fig. 9. Superposition of export gate complexes with YscVXY.
YscV, YscX, and YscY are shown in orange, cyan, and red, respectively. a Superposition with the cryo-EM structure of YscVC (gray, PDB: 7ALW), which is shown as cartoon representation. YscVC protomers are aligned with an r.m.s.d. of 0.797 Å. b, c Superposition with FlhA (gray) bound to the flagellar chaperone:substrate complexes (green:light green). b FlhA:FliS:FliC (PDB: 6CH3). c FlhA:FliT:FliD (PDB: 6CH2). Alignments were obtained using SD3 of YscVC (residues 514–600) with an r.m.s.d. of 1.194 Å (PDB: 6CH2) and 1.207 Å (PDB: 6CH3). d YscVXY superimposed with Chlamydia pneumoniae CdsV in complex with the stalk protein CdsO (PDB: 6WA9). The alignment was calculated using SD3 of YscVC with an r.m.s.d. of 0.854 Å. Two subunits of YscV and CdsV (gray) are shown, CdsO is colored in green.