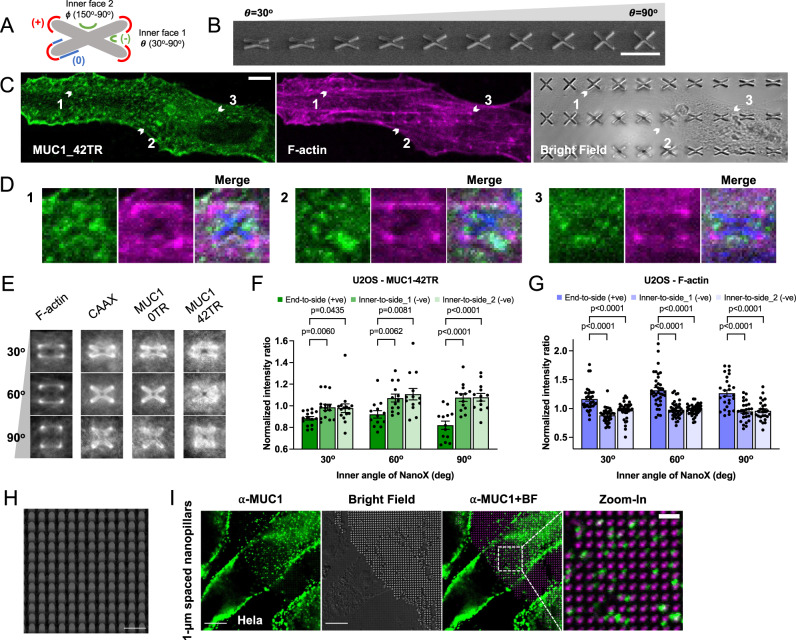
Fig. 2. MUC1 prefers negatively-curved over positively-curved membranes on the same nanostructures.
A Schematic illustration of a nanoX used to induce positive, negative, and zero membrane curvature on the same structure. B An SEM image of gradient nanoX arrays with inner angles ranging from 30° (left) to 90° (right), tilted at 45°. All nanoX are 350 nm in arm width, 5 µm in arm length, and 10 µm in spacing. NanoX inner angle (θ) increment: 15°. C Confocal images of MUC1_42TR-GFP-transfected U2OS cells cultured on the gradient nanoX arrays. F-actin was stained with phalloidin as a reference. D Three sets of zoom-in confocal images show that F-actin prefers the arm ends of nanoXs while MUC1_42TR-GFP prefers the inner faces. Bright-field images of nanoX structures were converted into blue color for visualization purpose. E Averaged fluorescence images show the spatial distributions of, F-actin, mCherry-CAAX, MUC1_0TR-GFP, and MUC1_42TR-GFP on nanoXs. F, G Quantification of end-to-side ratios (reflecting the preference for positive curvature) and inner-to-side ratios (reflecting the preference for negative curvature) of MUC1_42TR-GFP (F) and F-actin (G) on nanoXs of selected three inner angles. All ratios are normalized against the mCherry-CAAX signals (see Supplementary Table 3A, C for the detailed statistics). H An SEM image of a dense nanopillar array at 500-nm-diameter, 1-μm-height, and 1-μm-spacing with a stage tilt of 45°. I Confocal images of Hela cells cultured on the dense nanopillar array. MUC1 preferentially locates to inter-pillar spaces where negative membrane curvature can be induced. The bright-field (BF) channel in the merged image is background-subtracted and converted into magenta color for visualization. Scale bars = 10 µm for B, C, I. Scale bars = 2 µm for H and zoom-in images in I. Welch’s t-tests (unpaired, two-tailed, not assuming equal variance) are applied for all statistical analyses in this figure. Error bars represent SEM.