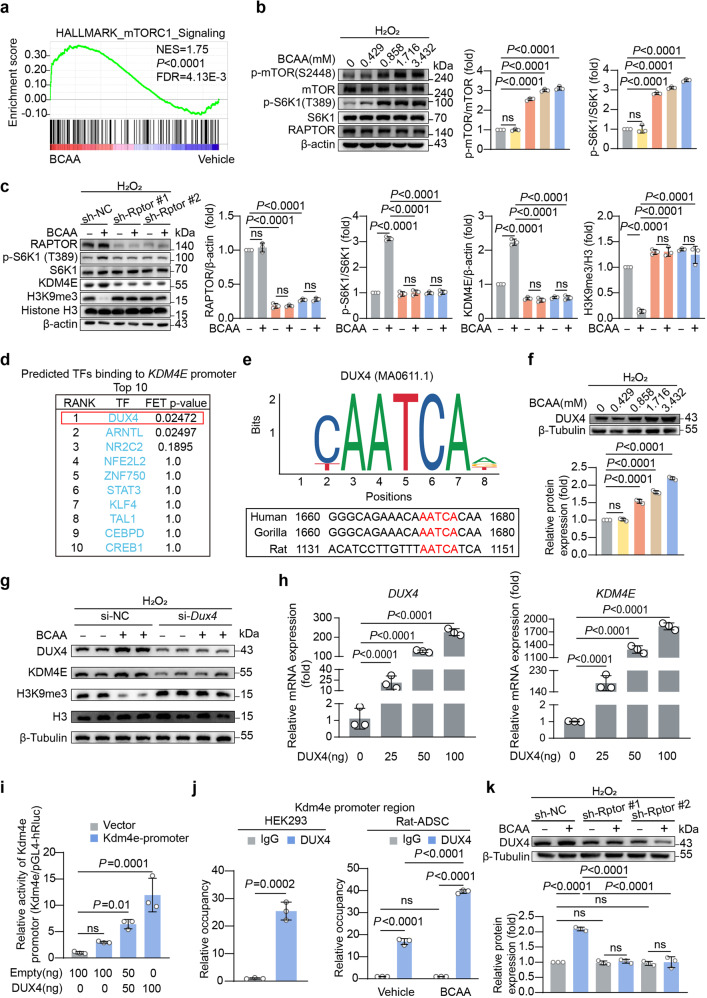
Fig. 5.
A novel mTORC1/DUX4/KDM4E axis was critical for BCAA-mediated H3K9me3 loss in ADSCs. a GSEA showed that the mTORC1 signaling pathway was significantly enriched in BCAA-treated ADSCs. b Representative blots of p-mTOR (S2448), mTOR, p-S6K1 (T389), S6K1, RAPTOR, and β-actin protein in ADSCs treated with increasing doses of BCAA in the presence of hydrogen peroxide (100 μM). c ADSCs were transfected with sh-Rptor#1, sh-Rptor#2, or sh-NC and treated with or without BCAA (3.432 mM) in the presence of hydrogen peroxide (100 μM). Representative blots of RAPTOR, p-S6K1 (T398), S6K1, KDM4E, H3K9me3, histone H3, and β-actin are shown. d Predicted TFs binding to the Kdm4e gene promoter region. The promoter region was defined as the 2000 bp upstream of the transcription start site of the Kdm4e gene. e Binding motif of DUX4 and its predicted binding site in the Kdm4e gene promoter region. f Representative blots and quantification of DUX4 protein in ADSCs treated with different doses of BCAA in the presence of hydrogen peroxide (100 μM). g ADSCs were transfected with si-NC or si-Dux4 and treated with or without BCAA (3.432 mM) in the presence of hydrogen peroxide (100 μM). Representative blots of DUX4, KDM4E, H3K9me3, H3, and β-tubulin are shown. h mRNA levels of Dux4 and Kdm4e in HEK293 cells transfected with plasmids overexpressing Dux4. Actb was used as the endogenous control gene. i Dual-luciferase activity assay. Vectors overexpressing Dux4 or empty vectors were co-transfected with the firefly luciferase reporter driven by the Kdm4e promoter in HEK293 cells. Renilla luciferase activity was used as a transfection control. j Enrichment of DUX4 protein within the promoter regions of the Kdm4e gene in HEK293 cells and ADSCs as determined by ChIP-qPCR. k Representative blots and quantification of DUX4 protein in sh-NC-, sh-Rptor#1-, and sh-Rptor#2-transfected ADSCs with or without BCAA treatment (3.432 mM) for 24 h. The data are shown as the means ± SD. The data shown in b, e, g, i, and j were analysed by one-way ANOVA followed by Bonferroni post hoc test. The data shown in c, g, and k were analysed by two-way ANOVA followed by a Bonferroni post hoc test. The data shown in j was analysed by unpaired Student’s t test. ADSCs adipose tissue-derived mesenchymal stem cells, BCAA branched chain amino acids, DUX4 double homeobox protein 4, H3K9me3 histone H3K9 trimethylation, KDM4E lysine-specific demethylase 4E, mTORC1 mechanistic target of rapamycin complex 1, S6K1 ribosomal protein S6 kinase polypeptide 1