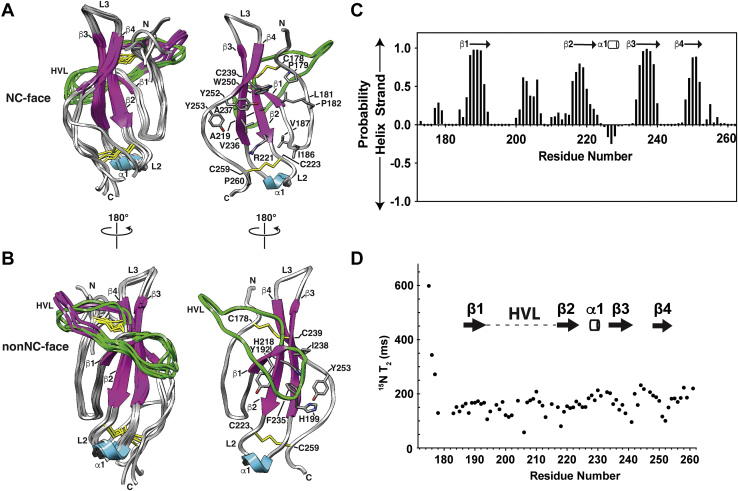
Figure 5.
Structure and backbone dynamics of TGM-D3.A and B, shown on the left are an ensemble of the five lowest-energy NMR structures of the unbound form of TGM-D3: β-strands, magenta; loops, gray; 310 helix, cyan; disulfide bonds, yellow, two conformations of HVL highlighted in green and pink. Key structural features are indicated. Orientations shown differ by a 180-degree rotation around the y-axis, with orientation shown in (A) highlighting the face of the protein that includes N- and C-terminus (NC-face) and the orientation shown in (B) highlighting the opposite face (non-NC-face). Shown on the right are single representative structures, with the four cysteines that form the two disulfide bonds and the side chains of key residues highlighted. C, PECAN-based prediction of TGM-D3 secondary structure. Positive values indicate β-strand probability; negative values indicate helical probability. Spectra recorded in 25 mM sodium phosphate, 50 mM sodium chloride, and 5% 2H2O, pH 6.0, at 310 K. Secondary structure elements shown above correspond to those deduced from the calculated TGM-D3 solution structure. D, backbone 15N T2 relaxation times for TGM-D3 plotted per individual residue with structural features mapped. HVL, hypervariable loop.