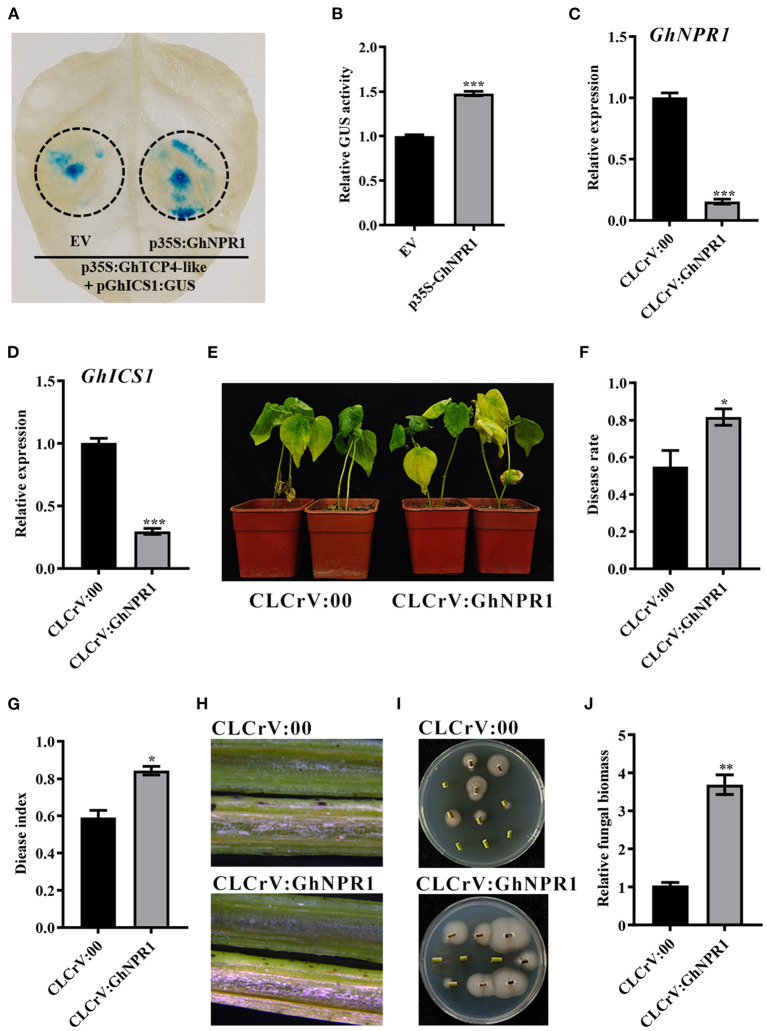
Figure 7.
GhNPR1 promotes transcriptional activating activity of GhTCP4-like for GhICS1 expression. (A) GUS staining assay. EV represents pCAMBIA1300. Tobacco leaves were harvested at 48 h after corresponding treatments. (B) GUS enzyme activity measurement. Error bar means SD of three independent biological replications. Student's t-test was performed, ***p < 0.001. (C) GhNPR1 expression level in CLCrV:00 and CLCrV:GhNPR1 plants. Total RNA was extracted 14 d after agroinfiltration. Error bar means SD of three independent biological replications. Student's t-test was performed, *p < 0.05. (D) Expression of GhICS1 in CLCrV:00 and CLCrV:GhNPR1 plants 1 dpi. Error bar means SD of three independent biological replications. Student's t-test was performed, ***p < 0.001. (E) Disease symptoms of CLCrV:00 and CLCrV:GhNPR1 plants after V. dahliae infection. Photos were taken at 21 dpi. (F,G) Disease rate and disease index of CLCrV:00 and CLCrV:GhNPR1 plants 21 dpi. Error bar means SD of three independent biological replications. Student's t-test was performed, *p < 0.05. (H) Color intensity of longitudinal sections of the stem of CLCrV:00 and CLCrV:GhNPR1 plants 21 dpi. Photos were taken by the body microscope. (I) Fungus discovery culture assay. Stems from CLCrV:00 and CLCrV:GhNPR1 plants 21 dpi were sprayed on PDA medium. Photos were taken after 5 d of culture at 25°C. (J) Relative fungal biomass in CLCrV:00 and CLCrV:GhNPR1 plants were determined by qPCR at 21 dpi. Error bar means SD of three independent biological replications. Student's t-test was performed, **p < 0.01.