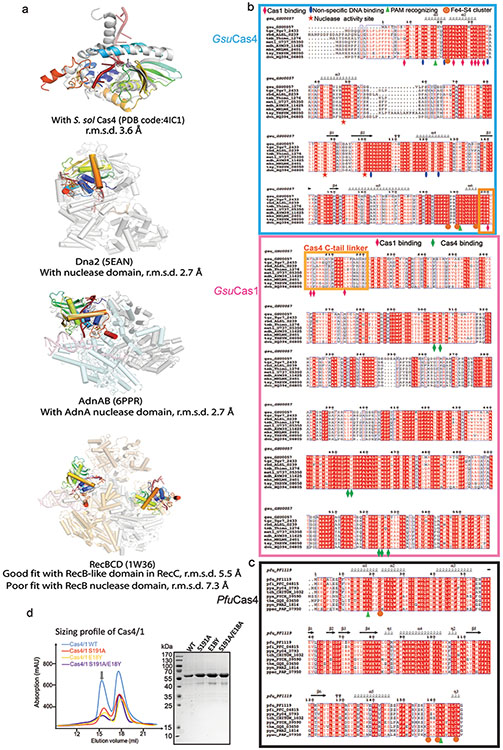
Extended Data Figure 5. In-depth analysis of the structure and sequence conservation in Cas4.
a. Superposition of GsuCas4 with a standalone Cas419,20, and the nuclease domains in helicase-nuclease fusion proteins AddAB32, AdnAB26, RecBCD33, and eukaryotic Dna234. The caging of the ssDNA substrate and the arrangement of the Fe-S cluster and the catalytic triad are conserved themes. Interestingly, the Cas4 structure aligns poorly with the RecB nuclease in RecBCD; it agrees better with the RecB-like fold in RecC instead, b, c. Sequence alignment of GsuCas4, GsuCas1, and PfuCas4 with their close homologs. Based on the structural analysis, we marked the residues important for subunit interaction, substrate binding, catalysis and Fe-S cluster formation, d. Quality of the purified GsuCas4 mutants that carry the PAM-recognition residues from PfuCas4. These mutants were used in the structure-guided PAM-switching experiments in Extended Data Fig. 4.