Abstract
Metalloenzymes catalyze a diverse set of challenging chemical reactions that are essential for life. These metalloenzymes rely on a wide range of metallocofactors, from single metal ions to complicated metallic clusters. Incorporation of metal ions and metallocofactors into apo-proteins often requires the assistance of proteins known as metallochaperones. Nucleoside triphosphate hydrolases (NTPases) are one important class of metallochaperones and are found widely distributed throughout the domains of life. These proteins use the binding and hydrolysis of nucleoside triphosphates, either adenosine triphosphate or guanosine triphosphate, to carry out highly specific and regulated roles in the process of metalloenzyme maturation. Here, we review recent literature on NTPase metallochaperones and describe the current mechanistic proposals and available structural data. By using representative examples from each type of NTPase, we also illustrate the challenges in studying these complicated systems. We highlight open questions in the field and suggest future directions. This minireview is part of a special collection of articles in memory of Professor Deborah Zamble, a leader in the field of nickel biochemistry.
Keywords: metallochaperones, GTPases, ATPases, metalloenzyme maturation, metallocofactors, metals in biology
Graphical Abstract
Graphical Abstract.
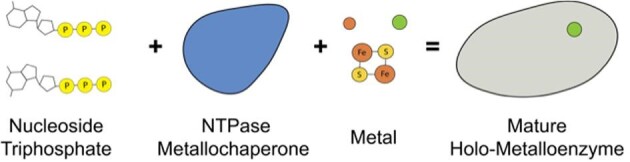
Nucleoside triphosphates are hydrolyzed by metallochaperones (blue) to insert metal ions, multi-metal clusters, or organometallic clusters to form the matured metalloenzyme (gray).
Introduction
Metal sites exist in about one-third of all structurally characterized enzymes, and over half of all proteins are predicted to be metalloproteins.1 These metal sites range in complexity from one metal ion to multi-metal or even organometallic clusters (Fig. 1).2 Metalloenzymes tend to catalyze the most challenging chemical transformations. Their roles include, but are not limited to, respiration3; photosynthesis4; regulation of transcription and translation3; and nitrogen,5 carbon,6 and hydrogen fixation.7 To have a functioning metalloenzyme, the metallocofactor must be correctly installed, which often requires a metallochaperone. Metallochaperones are proteins that physically interact with apo-metalloprotein clients or intermediary proteins to assist in cofactor delivery or assembly as part of the process of metalloprotein maturation. The exact percentage of metalloproteins that require metallochaperones for cofactor biogenesis is not established. Quantification is complicated by the fact that metallochaperones are often not needed for metalloprotein reconstitution in vitro but are in vivo, where metal ion concentrations are often limiting due to toxicity issues. In the cell, metallochaperones balance the cellular demand for metal-assisted reactivity with the toxicity associated with having too much metal.
Fig. 1.
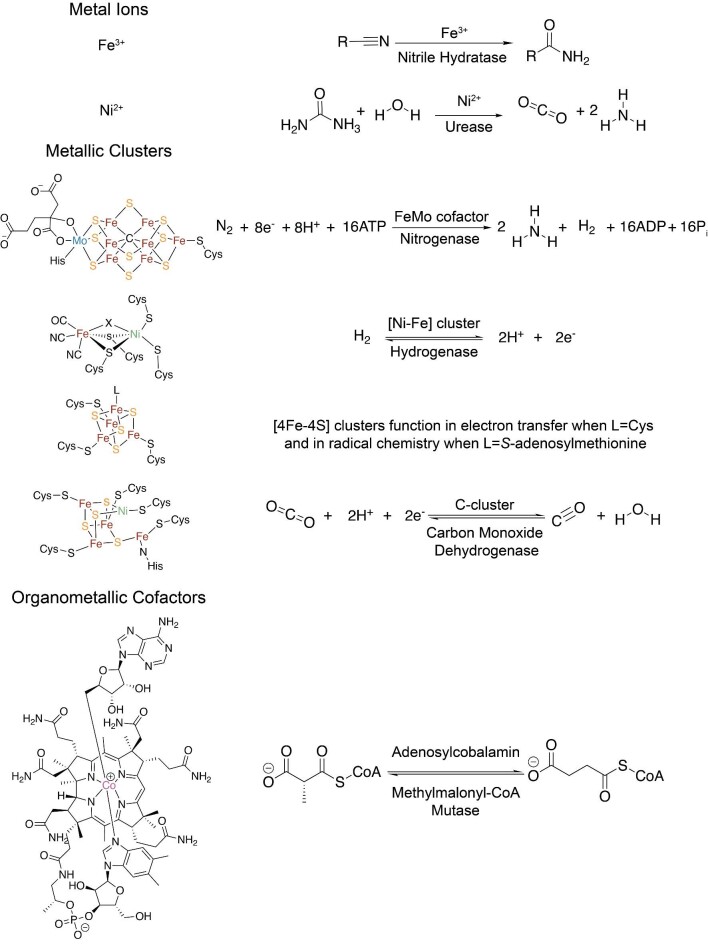
Metallocofactors vary widely in structure and reactivity. In some reactions, metal ions such as Fe3+ or Ni2+ are ligated by residues of the metalloprotein but still require metallochaperones for proper maturation. Increasingly complex metallic clusters, such as the FeMo-cofactor and [4Fe–4S] clusters, are transported and inserted by metallochaperones, whereas other metallic clusters, such as the [NiFe] cluster, are assembled in situ. Additionally, organometallic cofactors are often metabolically expensive and are transported by metallochaperones into their target metalloenzyme to perform difficult chemistry.
Deletion or mutation of genes encoding metallochaperones can lead to deleterious biological consequences. In Azobacter vinelandii, deletion of the metallochaperones NifX and NafY decreases the amount of active nitrogenase by half, reducing levels of dinitrogen reduction and stunting organismal growth.9,10 In humans, the metabolic disorder methylmalonic aciduria is caused by mutations to or deletion of the gene for methylmalonic aciduria type A protein (MMAA), a metallochaperone required for the maturation of adenosylcobalamin-dependent methylmalonyl-CoA mutase (MCM). Without the adenosylcobalamin cofactor, methylmalonyl-CoA cannot be converted into succinyl-CoA, causing an accumulation of methylmalonic acid that alters the blood pH leading to disease.11,12 Because of the deleterious effects of metallochaperone impairment, bacterial metallochaperones are also potential drug targets. For example, the metallochaperone UreG is necessary for the maturation of the dinickel metal active site of urease, an enzyme that catalyzes the hydrolysis of urea to ultimately yield ammonia and carbon dioxide, providing the buffering capacity necessary for Heliobacter pylori to live in the stomach. As such, urease is central to H. pylori metabolism and virulence,13,14 making its metallochaperones potential drug targets.
The wide variety of metal centers employed by metalloproteins requires a diversity of metallochaperones. There are numerous types of metallochaperones that have been characterized, from the transporters of the FeMo—cofactor of nitrogenase15,16 to the copper metallochaperones involved in shuttling copper ions to the mitochondrial electron transport chain for cellular respiration.17 This minireview focuses on one group of metallochaperones: nucleoside triphosphate hydrolase (NTPase) metallochaperones. NTPase metallochaperones use the binding and/or hydrolysis of nucleoside triphosphates (either GTP or ATP) for metalloenzyme maturation. They exist widely in the domains of life,18 and although they are ubiquitous, their exact roles in maturation processes are often not well understood. Known and postulated roles include: metal ion or metallocofactor transport to an intermediary protein in the metalloprotein maturation process; metallocofactor binding and direct insertion into apo-target protein; and induction of a conformational change in the target protein to allow for cofactor delivery. Here, we focus on the recent advances in the study of NTPase metallochaperones, describing the current state of knowledge and highlighting open questions.
P-loop G3E GTPase metallochaperones
The first subclass of NTPase metallochaperones that we will consider are phosphate-binding loop (P-loop)-containing G3E GTPases (hydrolyzing guanosine triphosphate, GTP).18 The P-loop G3E GTPase family is named for a D-to-E substitution in the G3 motif compared to the canonical P-loop GTPase, Ras.18 The best characterized subfamilies of the P-loop G3E GTPase metallochaperone family are the urease metallochaperone UreG,19,20 the [NiFe]-hydrogenase metallochaperone HypB,21 and the cobalamin metallochaperone MeaB (MMAA in humans) subfamilies.22–24 In addition to UreG, HypB, and MeaB/MMAA, there is a large and diverse subfamily of G3E GTPases called the cluster of orthologous groups (COG) 0523 proteins (Table 1). COG0523 proteins are characterized by a conserved CxCC (C = Cys; x = any amino acid) motif that is implicated in high affinity metal binding.25,26 Known functions of COG0523 proteins include: zinc homeostasis (ZigA/ZagA);27 cobalamin cofactor biosynthesis (CobW);28–30 and nitrile hydratase maturation (Nha3).31
Table 1.
Subfamilies of the G3E P-Loop GTPases
| Subfamily | Known roles | Known metals/metallocofactor |
|---|---|---|
| UreG | Maturation of ureases | Nickel |
| HypB | Maturation of [NiFe]-hydrogenases | Nickel |
| MeaB | Maturation of adenosylcobalamin-dependent mutases | Cobalamin |
| COG0523 | Zinc homeostasis (ZigA/ZagA), Nitrile hydratase maturation (Nha3), Cobalamin cofactor biosynthesis (CobW) |
Zinc Iron Cobalt |
P-loop G3E GTPases employ a common protein fold
All the known structures of the G3E P-loop GTPases contain a G-domain (Table 2) that is comprised of a very common protein fold32: regularly recurring α–β units with the β strands forming a central β-sheet surrounded on both sides by α-helices (Fig. 2). The typical G-domain contains five different conserved motifs, known as G1–G5, that are involved in nucleotide binding and hydrolysis (Fig. 2). The G1 motif, also known as the Walker A motif, is a flexible loop in between a helix and a sheet. This motif functions to position the triphosphate of the bound nucleotide. The G2 motif, also known as switch I, signals which nucleotide, if any, is bound. The G3 motif, also known as the Walker B motif loop, is often situated at the end of a strand and contains a conserved aspartate or, less commonly, glutamate residue that binds the water-bridged Mg2+ ion used for NTP hydrolysis.32 The G4 motif typically contains the sequence motif NKXD that is used in the recognition of the guanosine base.18 Finally, the G5 motif is involved in nucleotide release.32 The ubiquitous nature of P-loop NTPases has led to further classification of various structural motifs that are beyond the scope of this review but have been previously detailed by Leipe et al.18
Table 2.
Example structures of members of the G3E P-loop GTPases
| Subfamily | Protein | Organism | PDB IDs: ligand(s) bound |
|---|---|---|---|
| MeaB | MeaB | Methylobacterium extorquens | 2QM8: no ligands bound33 2QM7: GDPa bound33 4JYB: GMPPNPb bound34 |
| MMAA | Homo sapiens | 2WWW: GDP bound35 | |
| MeaB fusion protein (IcmF) | Cupriavidus metallidurans | 4XC7: no ligands bound36 4XC8: GDP and Mg2+ bound36 |
|
| HypB | HypB | Methanocaldococcus jannaschii | 2HF9: GTPγSc and Mg2+ bound37 |
| Heliobacter pylori | 4LPS: GDP, Mg2+, Ni2+ bound38 | ||
| UreG | UreG | Klebsiella pneumoniae | 5XKT: GMPPNP, Ni2+ bound39 |
| UreG/UreF/UreH | Helicobacter pylori | 4HI0: GDP bound14 | |
| COG0523 | YjiA | Escherichia coli | 4IXM: Zn2+ bound26 |
aGDP: guanosine diphosphate.
bGMPPNP: Guanosine 5′-[β,γ-imido]triphosphate.
cGTPγS: guanosine 5′-O-(3-thiotriphosphate).
Fig. 2.
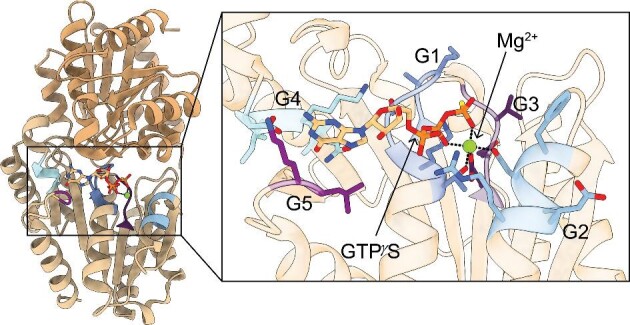
Dimeric structure of P-loop-containing G3E GTPase metallochaperone with conserved motifs. (Left) Representative structure: HypB from Methanocaldococcus jannaschii (PDB 2HF8) with guanosine 5′-O-(3-thiotriphosphate) (GTPγS) and Mg2+ bound.37 Structure is a dimer with each monomer containing a G-domain. (Insert) The GTP-binding site contains the G1–G5 motifs, which are highlighted. The G1 residues (dark blue), also known as the Walker A motif, interact with the α and β phosphates and the bound Mg2+ ion. The G2 residues (light blue), also known as switch I, change conformation based on the nucleotide state and the conserved aspartate residue coordinates the bound Mg2+ ion. The G3 residues (dark purple), also known as the Walker B motif, coordinate the γ-phosphate and the bound Mg2+ ion. The G4 residues (cyan) confer nucleotide specificity as they interact with the base of the nucleotide. The G5 residues (light purple) are involved in nucleotide dissociation.
The mechanism of GTP hydrolysis is well established
The mechanism of nucleoside triphosphate hydrolysis by various GTPases has been extensively studied.32,40–43 Briefly, after nucleotide triphosphate binding, the active site is completed by a Mg2+-bound water molecule positioned by an aspartate (or less commonly, glutamate) to perform the hydrolysis of the terminal phosphate moiety of the nucleoside triphosphate. After the cleavage of the bond to the γ phosphate, the resulting nucleotide is a diphosphate; when replaced with a nucleoside triphosphate, the cycle can proceed again. Oftentimes, this binding and/or cleavage occurs in the presence of another protein known as an activating factor that increases the intrinsic rate of hydrolysis of the GTPase. In the case of metalloprotein maturation, the activating factor can be the target enzyme or another protein involved in the maturation process. Similarly, a nucleotide exchange factor is sometimes needed to replace the nucleoside diphosphate with the corresponding triphosphate.32 The changes throughout this cycle form the basis for the ability of the GTPases to perform their chaperone roles in the maturation of metalloproteinases.
The well-studied family members UreG, HypB, and MeaB show mechanistic diversity
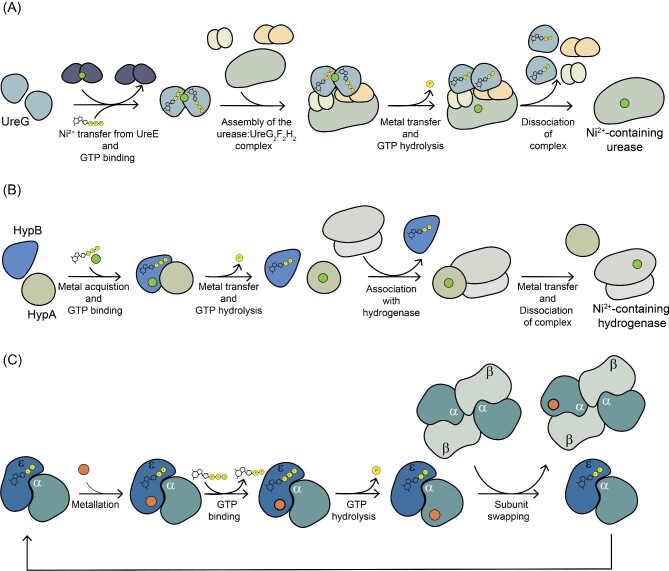
The most well-studied family members of the G3E P-loop GTPase metallochaperones utilize different mechanisms for maturing their respective metalloenzymes despite employing a common, and potentially ancient protein fold.18 As far as we know, the evolutionary factors that drove these mechanistic differences within this metallochaperone subfamily are not understood. These mechanistic differences are especially interesting given that both UreG and HypB are metallochaperones involved in the maturation of the nickel-dependent enzymes: urease and [NiFe]-hydrogenase, respectively. UreG undergoes a conformational change when it binds GTP and accepts Ni2+ from UreE. Then, UreG forms a complex with apo-urease and the other accessory factors UreF, UreH, and UreD to directly insert the Ni2+ into its target enzyme, urease, through a tunnel created by the complex of all the accessory factors (Fig. 3A).14,39,44 Although HypB is also conformationally gated by GTP binding and hydrolysis, it does not directly insert Ni2+ into the [NiFe]-hydrogenase active site. The Zamble Lab showed that it is the GDP-loaded state of Escherichia coli HypB that is “readied” for fast Ni2+ transfer to accessory protein HypA, which ultimately leads to Ni2+ insertion into the active site of [NiFe]-hydrogenase (Fig. 3B).45 However, not all GTPase metallochaperones directly bind the metallocofactor that they are involved in delivering. In the maturation of methylmalonyl-CoA mutase, the GTPase metallochaperone MeaB (in bacterial systems, MMAA in humans) is required for adenosylcobalamin delivery; however, there is no evidence of the chaperone interacting with the cofactor itself.24,35,46–49
Fig. 3.
Postulated roles of G3E P-loop GTPases in metalloenzyme maturation. (A) Urease (pale green) requires the GTPase metallochaperone UreG (light blue) for insertion of the required Ni2+ ion. UreG receives the Ni2+ ion from UreE (dark blue) and binds GTP. The GTP binding event allows for the assembly of the apo-urease: the UreG2F2H2 complex. GTP hydrolysis triggers a conformational change that facilitates metal transfer to urease. After GTP hydrolysis occurs, the complex dissociates, and the urease now contains the needed Ni2+ ion for a fully activated enzyme.14,19,39 (B) [NiFe]-hydrogenases (light gray) require the metallochaperone HypB (blue) for Ni2+ ion insertion. HypB binds GTP and acquires Ni2+, which is transferred to HypA (pale green). HypA then interacts with the inactive hydrogenase to transfer the metal to the active site of the [NiFe]-hydrogenases for activation.21,44,45,93,99 (C) Fe-type NHases maturation with Fe ions (orange)51,52 involves a COG0532 GTPase metallochaperone, called ε or Nha3 (blue), which forms a complex with the α subunit of NHase (gray). The holo α subunit of NHase is swapped for an apo α subunit of NHase from the NHase α2 β2 complex. The process is repeated to fully mature the Fe-type NHases using the activating protein Nha3.51
COG0523 subfamily and its best characterized member Nha3
Giedroc and coworkers recently provided a comprehensive comparative phylogenetic, biochemical, structural, and functional analysis of P-loop G3E GTPases in the COG0523 subgroup.50 These proteins typically have two domains, a conserved G-domain at the N-terminus and a variable C-terminal domain that is predicted to be involved in target-specific protein interactions. The sequence analysis by Giedroc et al. suggests that there are multiple subfamilies within the COG0523 subgroup and that there is a considerable amount of unexplored sequence space. Many of the COG0523 sequence clusters have no characterized members. The lack of functional data is especially notable for COG0523 proteins from eukaryotic organisms. The best characterized C0G053 member is Nha3, a Fe-type nitrile hydratase (NHase) maturase. Currently, it serves as the model system for this diverse GTPase metallochaperone protein family.
Although Co-type NHases do not appear to require a GTPase metallochaperone,31 Fe-type NHases do. NHases catalyze the hydrolysis of organic nitrile to the corresponding amide product (Fig. 1) by using a metal ion (Co3+ or Fe3+) that is buried at the interface between the α and β subunits.51,52 The maturation of Co-type nitrile hydratases relies on an α subunit swapping mechanism that requires a GTP-independent maturation protein,31 whereas the maturation protein for Fe-type NHases is a COG0532 GTPase metallochaperone (called ε or Nha3) (Fig. 3C). Briefly, the swapping mechanism for the Fe-type NHases appears to involve a Fe2+-loaded GTP-bound Nha3 forming a complex with an apo α subunit of Fe-type NHase.51,52 In a GTP-dependent manner, the α subunit receives the metal and is oxidized to Fe3+ in the α subunit. Consistent with other G3E P-loop GTPases, the presence of divalent metals bound increases the rate of GTP hydrolysis of Nha3.51 The apo complex of the α2β2 NHase swaps an apo-α subunit with a holo-α subunit to form the holo-NHase.51 Substitutions of the conserved Lys or Thr of the P-loop of Nha3 from Pseudomonas chlororaphis B23 result in an in vivo loss of detectable NHase activity similar to what has been observed for UreG and urease and HypB and [NiFe]-hydrogenase, further implicating the GTPase activity of Nha3 in Fe-type NHase maturation.53
ATPase metallotransporters and metallochaperones
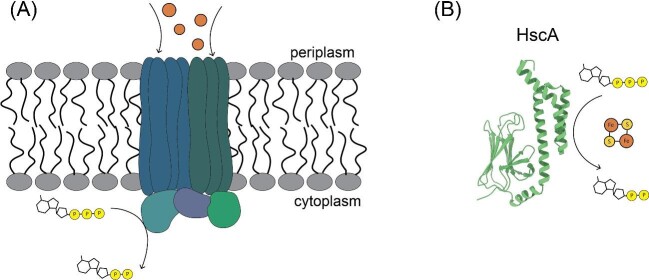
Compared to GTPases, there is a wider variety in the types of characterized ATPases (hydrolysis of adenosine triphosphate, ATP) involved in metalloprotein maturation (Fig. 4, Table 3).54,55 The P1B class of P-type ATPases uses the energy of ATP hydrolysis to transport d-block metal ions across cell membranes. In plants, the P1B class of P-type ATPases are key components in the maintenance of metal homeostasis, with characterized transporters for Zn2+, Cu2+, Cd2+, Co2+, and Pb2+ ions.54 These transporters serve as the first step in metalloprotein maturation by acquiring d-block metal ions from the surrounding environment. Analogous to the P-loop GTPases described above, P-loop ATPases are also involved in transporting metal ions to the active site of their target protein. One well-characterized P-loop ATPase metallochaperone is CooC,56 which is involved in the maturation of carbon monoxide dehydrogenases (CODHs).57 CODHs catalyze the reversible oxidation of carbon monoxide at a metallocofactor, the C-cluster, which consists of Ni, Fe, and S (Fig. 1). CooC is responsible for ATP-dependent Ni2+ insertion as part of the process of C-cluster maturation.56–58 Additionally, ATPases containing a heat shock protein fold are known to be involved in the biogenesis and transport of various metallic clusters. For example, in the iron-sulfur cluster (ISC) pathway found in mammals, the ATPase HscA works with the co-chaperone HscB to stimulate the transfer of nascently synthesized [2Fe–2S] to various apo enzymes in an ATP-dependent manner.55,59
Fig. 4.
Some representative ATPases involved in metalloprotein maturation. (A) P1B-type ATPases are involved in the transport of heavy metals (orange spheres) into the cells to maintain metal homeostasis. The protein contains two domains: a transmembrane domain and a cytoplasmic ATPase domain. The flux of heavy metals is coupled to the hydrolysis of ATP.54 (B) The heat shock protein fold ATPases, such as HscA from E. coli (PDB 1U00), interact with co-chaperones in a nucleotide-dependent manner to transfer newly synthesized [2Fe-2S] clusters in the ISC biogenesis pathway.100
Table 3.
Subtypes of ATPase metallochaperones
| Subtype | Known roles | Known metals/metallocofactor |
|---|---|---|
| P1B class | Transporting metal ions across membranes | d-block metals |
| P-loop | Maturation of metalloenzyme active sites | Ni2+ |
| Heat shock fold proteins | Biogenesis of Fe–S clusters | Fe–S clusters |
Structures of P-loop ATPase CooC are similar to P-loop GTPase HypB
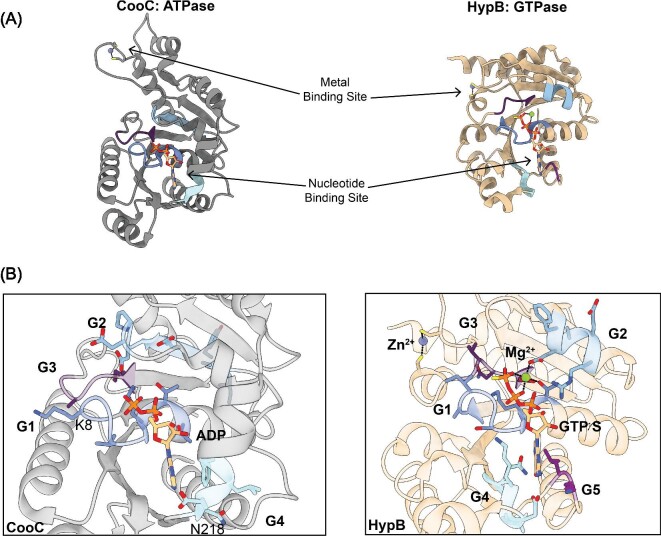
Comparisons of the structures of CooC, a P-loop ATPase, and HypB, a P-loop GTPase, reveal an incredibly similar overall architecture of α–β units, despite low sequence homology of around 25% (Fig. 5A). Both proteins contain the G1–G3 sequence motifs that are involved in binding the phosphates of the nucleotide and the Mg2+ ion, although CooC contains the deviant G1 motif with the highly conserved lysine.60 The deviant G1 motif has the highly conserved residue of a typical G1 motif in the second position instead of the second-to-last position in the sequence motif, which is a characteristic of the MinD/BioD class of P-loop NTPases.18 The G4 nucleotide specificity motif in CooC is different from the conserved NKXD motif of HypB in that it only retains the first asparagine, and the CooC adenine base interacts with the residues of an alpha helix that is ordered upon binding the nucleotide (Fig. 5B).60 Additionally, these two proteins both have spatially separated but nucleotide-coupled metal-binding sites that control the oligomeric state of the NTPases (Fig. 5A).60
Fig. 5.
Comparison of the ATPase CooC and the GTPase HypB. (A) The ATPase metallochaperone CooC from C. hydrogenoformans (PBD 3KJI) (left, gray)60 and the GTPase metallochaperone HypB from M. jannaschii (PBD 2HF8) (right, tan)37 contain the conserved overall fold found in P-loop NTPases consisting of repeating α–β units. The metal-binding site and the nucleotide-binding site are spatially separated but involved in controlling the oligomeric states of the NTPases. Only one monomer for each metallochaperone is shown for simplicity. (B) (Left) A closer view of the ATP-binding site of CooC bound to ADP indicates that it contains the conserved G1–G3 motifs implicated in interacting with the phosphates of the nucleotide and the Mg2+ ion if present. The G1 residues (dark blue), also known as the Walker A motif, interact with the α and β phosphates and the bound Mg2+ ion. CooC contains the deviant Walker A motif with a highly conserved lysine residue (K8 in CooC) in the second position of the sequence instead of the second to last position. The G2 residues (light blue), also known as switch I, change conformation based on the nucleotide state and the conserved aspartate residue coordinates the bound Mg2+ ion. The G3 residues (dark purple), also known as the Walker B motif, coordinate the γ-phosphate and the Mg2+ ion if present. CooC only retains the first asparagine residue (N218 in CooC) of the G4 motif (cyan) that confers nucleotide specificity. An alpha helix, which is only present when a nucleotide is bound, interacts with the adenine base. (Right) A closer view of the GTP-binding site of HypB bound to GTPγS and a Mg2+ ion reveals the canonical G1–G5 motifs described in Fig. 2. Unlike CooC, HypB contains the conserved G4 motif (cyan) that confers nucleotide specificity by interacting with the guanosine base. The G5 residues (light purple) are involved in nucleotide dissociation and are not conserved in CooC.
Fe–S cluster biogenesis utilizes ATPases
Fe–S cluster biogenesis is a highly conserved yet complicated process that involves dedicated machinery to synthesize, transport, and deliver Fe–S clusters. This dedicated machinery involves proteins that act as chaperones and/or “scaffolds” for the assembly and delivery of Fe–S clusters. Notably, many of these proteins have ATP-binding sites and/or ATPase activity. There are four main Fe–S cluster biogenesis pathways: the sulfur fixation (SUF), the ISC, the nitrogen fixation (NIF), and the cytosolic iron-sulfur cluster assembly (CIA) systems. The best characterized Fe–S cluster biogenesis pathway is the ISC system, which requires the HscA/HscB [also known as heat shock protein 70 (Hsp70)/J-protein] chaperone/co-chaperone ATPase complex. Like most NTPases, these chaperone/co-chaperone proteins use nucleoside triphosphate binding and hydrolysis to regulate the transfer of a newly synthesized Fe–S cluster from a scaffold protein to an apo-recipient.61,62 The NIF system uses analogous chaperone/co-chaperone proteins but also requires an ATPase to deliver homocitrate-bound molybdenum during the process of assembling the FeMo-cofactor of nitrogenase; the roles of ATPases in nitrogenase maturation have been recently reviewed.5,63 In the SUF pathway, the ATP-binding cassette (ABC)-type ATPase SufC is necessary for Fe–S cluster formation.64 SufB, which accepts sulfur, and SufD, which has been proposed to play a role in iron acquisition, have been shown to interact with each other in vitro.64,65 Along with SufB and SufD, deletions of SufC abolish SUF function in vivo. In vitro, the SufBC2D complex can serve as a scaffold for de novo Fe–S cluster biogenesis.66 Recent structural studies of the SufBC2D scaffold provide insight into how ATP binding to SufC may promote conformational changes that are necessary for the formation of the cluster assembly site; however, a detailed mechanistic understanding of the ATPase cycle for cluster formation is not yet available.64,67 Finally, the CIA pathway uses a scaffold ATPase to assemble new Fe–S clusters; ATPase activity is required for both [2Fe–2S] cluster acquisition and transfer of the fully formed [4Fe–4S] cluster to the apo-client in the cytosol or nucleus.68–70 Although many of the participants of these pathways have been identified, their biochemical characterizations are limited.62 Recently, Perlstein and co-workers have provided a biochemical roadmap for exploring the roles of ATP-binding sites and ATPase activity for proteins involved in Fe–S cluster biogenesis.71
Nucleotide specificity of NTPase metallochaperones can be challenging to establish
Not all NTPase metallochaperones fall clearly into a GTPase subgroup or an ATPase subgroup. Although the better studied HypBs are GTPases, some organisms have versions of HypB that are ATPases.72,73 The HypB from the archaea Thermococcus kodakarenis is a P-loop ATPase containing the same overall fold as the characterized GTPase HypBs. Despite less biochemical characterization, there is evidence in some organisms that the HypB ATPases are functional homologs that replace the HypB GTPases, with their activity also regulated by Ni2+-binding, ATP hydrolysis, and interactions with HypA.73 Also, the Feo iron transport system appears to utilize an NTPase called FeoB for which the nucleotide specificity has been unclear.74–80 The Feo system is a dedicated Fe2+ ion transport system that is widely present in the archaeal and bacterial domains of life. It is composed of FeoA and FeoC, cytosolic proteins, and the transmembrane iron permease, FeoB. FeoB from the human pathogen H. pylori is thought to be an ATPase due to impaired Fe2+ ion transport when whole cells are treated with known inhibitors of ATP synthesis or hydrolysis.81 In contrast, FeoB from E. coli is believed to be a GTPase because no ATP binding was observed in vitro.82 The characterization of the FeoB from Vibrio cholerae showed both low intrinsic GTPase and ATPase activity that was not stimulated by any known factors.78,79 Research by Kim and coworkers on various pathogenic bacterial FeoBs suggests that there are two classes of FeoBs: sole GTPases and promiscuous NTPases.80
FeoB, and also HypB, highlight the difficulty in establishing nucleotide specificity for NTPase metallochaperones. It is clear in the case of HypB, and likely in the case of FeoB, that different organisms employ different NTPases (GTPases or ATPases). The reason(s) for this variation are not well established. However, it also seems to be the case that some metallochaperone systems have no variation; for example, there are no known MeaB metallochaperones that are ATPases. Yet in other systems, NTPases appear to be employed that are promiscuous. It is not known if this promiscuity is an in vitro or in vivo feature. Given that most NTPase metallochaperones are poor NTP hydrolases without their target protein and/or without the appropriate metal ion bound, one must be careful about drawing conclusions, both favorable and unfavorable, from low levels of NTP hydrolysis activity. Additional studies may reveal that all NTPases are specific under the right set of conditions and/or that more GTPases have ATPases cousins. A MeaB that is an ATPase may be discovered, for example. It is currently not possible to predict nucleotide specificity from protein sequences, but more structural data will likely improve the prediction possibilities.
Metal specificity of NTPase metallochaperones can be challenging to establish
An important role of metallochaperones is to make sure that the correct metal ion is inserted into the correct apo-metalloprotein target, raising the question of how the metal specificity of metallochaperones is established.83 The biological challenge of correct protein metalation has been recently reviewed.84 Briefly, one important factor in metalation is cellular metal availability, which is thought to be opposite of the Irving–Williams series, Mg2+< Mn2+ < Fe2+ < Co2+ < Ni2+ <Cu2+ > Zn2+, with the weaker binding metals such as Mg2+, Mn2+, and Fe2+ more widely available and the tighter binding metals such as Ni2+, Zn2+, and Cu2+ less available.85–87 The reducing/oxidizing environment in the cell is also a consideration in terms of availability. For example, copper ions in the cell are thought to be in the Cu1+, rather than Cu2+ state given the reducing environment of the cytoplasm. Availability further depends on the ability of the organism to uptake metal ions, especially trace metals. In addition to availability, another factor affecting specificity is the metallochaperone's affinity for different metal ions, a feature of the protein that can be modulated by GTP binding or hydrolysis. All of this information points to the fact that establishing the metal specificity of a metallochaperone can be challenging.88
Establishing metal specificity for the GTPase metallochaperone CobW
The observation that cobW gene disruption in Pseudomonas denitifricans impairs aerobic cobalamin biosynthesis led to the proposal that CobW is a Co2+-dependent metallochaperone.29,30 To test this proposal, affinities were measured for Co2+ and other metal ions for the GTPase-dependent CobW from Rhodobacter capsulatus in the presence and absence of nucleotide effectors.89 Only weak interactions between CobW and Co2+ ions were observed in the absence of nucleotides. The addition of GTP or less hydrolyzable analogs promotes the tight coordination of two Co2+ ions in two different binding sites with different affinities. However, if Mg2+ ions are also present at physiological concentrations, the coordination of a Co2+ ion is observed in only one binding site. The other site is occupied by a Mg2+ ion. It is thought that the Mg2+ ion binds first in the weak affinity metal-binding site, ordering the second binding site for the high affinity binding of a Co2+ ion. When GDP is present instead of GTP, Co2+ binds CobW with a 1000-fold weaker affinity, indicating that an intact γ-phosphate is required for tight binding. The presence of bound GDP causes CobW to release the Co2+, further tying the nucleotide state to the metalation state of CobW. Finally, when compared to other first row transition metals, Mg2+GTP-CobW binds Zn2+ and Cu1+ more tightly;89 however, using the idealized pool of bioavailable metals90 and the free energy change for metal binding, calculations indicated that in vivo over 90% of Mg2+GTP-CobW would be bound with Co2+, as compared to less than 10% bound with Zn2+ due to the greater favorable free energy change for Co2+ binding.89 This recent work firmly establishes Co2+ as the cognate metal for CobW, supporting its known involvement in cobalamin biosynthesis. Additionally, these studies confirm the presumed role of GTP binding and hydrolysis for metalation of the metallochaperone, like other G3E P-loop GTPases.89
Final thoughts and future directions
The roles of NTPase metallochaperones continue to expand with new functions being established. However, there is much more work to be done. The COG0523 subfamily of G3E P-loop GTPases, for example, represents a poorly understood class of GTPase metallochaperones that are believed to bind and insert transition metals.50 Many of these putative GTPases are uncharacterized and their target client proteins are unknown. Bioinformatic studies of the gene clusters containing these uncharacterized GTPases may allow for predictions of the target client proteins. Predictions should be followed by biochemical characterizations exploring NTPase:target protein interactions, NTP specificity, and metal ion specificity. The latter can potentially be aided by the application of methods/calculations used in the CobW studies described earlier.89
Genomic initiatives and the wealth of sequence information that they generate are leading to proposals of putative NTPase metallochaperones outside of the COG0523 family. For example, the protein MutS is well known for its involvement in DNA repair processes;91 however, bioinformatics has revealed sequences of MutS-like proteins in operons associated with adenosylcobalamin-dependent enzymes, leading to a proposal that MutS variants might play a role in ATP-dependent metalloprotein complex assembly.92 We hypothesize that there are likely other examples of ATPase families whose distant cousins are involved in diverse biological processes. We are excited about how modern bioinformatics methods combined with genomic data will undoubtedly expand the NTPase metallochaperone field in the near future.
Among the already characterized NTPase metallochaperones, numerous questions remain. For example, it is often unclear whether the complete complement of stimulatory factors that increase NTP hydrolysis has been identified; is another protein involved or maybe another metal ion? Molecular mechanistic questions are also prevalent; what exactly is the function of the nucleotide-state-dependent conformational change, and what type of conformational change occurs? Structural information on GTPase metallochaperones is fairly limited (Table 2), and even when structural methods have allowed for the capture of more than one nucleotide-bound state, it is often the case that crystal lattice contacts have prevented the conformational change from occurring.36 Additionally, the absence of the target protein or other stimulatory factors may hinder the nucleotide-state-dependent conformational change of the NTPase from being fully realized.33 In other words, obtaining the requisite structural snapshots of NTPases to understand their molecular mechanisms is not trivial, and for the most part, these structural data are missing for NTPase metallochaperones, leaving molecular mechanistic questions unanswered. For example, there is more to learn about the steps required to transfer nickel from the GTPase HypB to its partner metallochaperone HypA to complete the [NiFe]-hydrogenase active site. The extent of the conformational changes that occur in HypB due to GTP binding and hydrolysis has not been visualized, limiting the molecular understanding of the mechanism.45,93 In the maturation of the adenosylcobalamin-dependent methylmalonyl-CoA mutase, the GTPase MMAA (MeaB in bacteria) is required for adenosylcobalamin insertion but does not bind the cofactor directly.24,35,46–49,94 Although there are a number of structures available of MMAA/MeaB in various nucleotide-bound states,33–35 and even a structure of a MeaB-fusion protein in which the chaperone domain is covalently attached to the target enzyme,36 the molecular basis by which MMAA/MeaB facilitates adenosylcobalamin delivery is still in debate. Currently, none of the structural rearrangements observed explain the molecular basis of methylmalonyl-CoA mutase maturation.34,36 Cryo-electron microscopy (cryo-EM) represents a promising new direction for the obtainment of these requisite structural data as this method allows for structures of protein:protein complexes to be obtained and for multiple conformations of proteins to be more readily visualized. Although the NTPases by themselves are too small for cryo-EM, the protein:protein complexes involved in metalloprotein maturation should be sufficiently large,95 and ultimately, it is the structures of protein:protein complexes that are needed for a molecular understanding. Thus, the resolution revolution of cryo-EM, i.e. the revolutionary ability to determine near-atomic resolution protein structures by cryo-EM, represents an exciting prospect moving forward for the determination of structures of metalloprotein maturation machineries.
An improved understanding of metalloenzyme maturation has several possible industrial applications. For example, nitrogenases are attractive as environmentally friendly alternatives to the industrial Haber–Bosch process, which is estimated to use ∼1% of the world's energy. This alternative solution would be even more attractive if nitrogenases could be prepared at high levels with their complex metallocofactors correctly inserted (Fig. 1). Hydrogenases are appealing for use in biofuel cells, and CODHs for fixation of the greenhouse gas carbon dioxide. Again, these applications require that the metalloproteins be produced in high yield, which necessitates an understanding of cofactor biogenesis/delivery. Additionally, knowledge of metalloenzyme maturation processes could be exploited to deliver synthetic metallocofactors with altered reactivity to apo target enzymes.
Regarding human health, understanding the molecular mechanisms of NTPase metallochaperones may provide novel solutions for therapeutics and treatments.11,17,96–98 For example, methylmalonic aciduria, an inborn error of metabolism, is caused by deletions of or mutations to any of the numerous proteins that transport and/or insert adenosylcobalamin into methylmalonyl-CoA mutase, including MMAA. Since MMAA does not interact with the cofactor directly, understanding how MMAA uses GTP binding and hydrolysis to perform its gatekeeping role could provide a novel therapeutic strategy. Additionally, NTPase metallochaperones thought to be involved in virulence, such as FeoB and UreG in H. pylori, are additional drug targets.13,81
In closing, we dedicate this minireview to the memory of Professor Deborah Zamble, who was a leader in the field of nickel enzymes and contributed to much of the research described earlier. Deborah left us too soon, and the work she started is not yet complete. There are many fascinating aspects of NTPase metallochaperones awaiting discovery and applications of metalloenzymes to be pursued. For young scientists looking to make a mark, metalloprotein maturation processes are a ripe area for discovery.
Acknowledgements
This work was supported by National Institutes of Health (NIH) grant R35 GM126982 to C.L.D. and an MIT Dean's Fellowship and an NIH F31 predoctoral fellowship GM131648 to F.A.V. C.L.D. is a Howard Hughes Medical Institute Investigator and Senior Fellow for the Canadian Institute for Advanced Research Bio-Inspired Solar Energy program.
Contributor Information
Francesca A Vaccaro, Department of Chemistry, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, MA, USA.
Catherine L Drennan, Department of Chemistry, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, MA, USA; Department of Biology, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, MA, USA; Howard Hughes Medical Institute, Massachusetts Institute of Technology, Cambridge, MA, USA.
Conflicts of Interest
The authors have no conflicts or competing interests to declare.
Data Availability
No new data were generated or analyzed in support of this research.
References
- 1. Degtyarenko K., Metalloproteins, In Jorde L. B., Little P. F. R., Dunn M. J., Subramaniam S., (ed.), Online Encyclopedia of Genetics, Genomics, Proteomics and Bioinformatics, Wiley, 2005. [Google Scholar]
- 2. Hausinger R. P., New metal cofactors and recent metallocofactor insights, Curr. Opin. Struct. Biol., 2019, 59, 1–8. [DOI] [PubMed] [Google Scholar]
- 3. Wachnowsky C., Fidai I., Cowan J. A., Iron-sulfur cluster biosynthesis and trafficking - impact on human disease conditions, Metallomics, 2018, 10 (1), 9–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yruela I., Transition metals in plant photosynthesis, Metallomics, 2013, 5 (9), 1090–1109. [DOI] [PubMed] [Google Scholar]
- 5. Lee C.-C., Stiebritz M. T., Hu Y., Ribbe M. W., Assembly and function of nitrogenase, In Moura J. J. G., Moura I., Maia L. B. (ed.), Enzymes for Solving Humankind's Problems: Natural and Artificial Systems in Health, Agriculture, Environment and Energy, Cham: Springer International Publishing, 2021, pp. 155–184. [Google Scholar]
- 6. Ragsdale S. W., Nickel and the carbon cycle, J. Inorg. Biochem., 2007, 101 (11–12), 1657–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shafaat H. S., Rüdiger O., Ogata H., Lubitz W., [NiFe] hydrogenases: a common active site for hydrogen metabolism under diverse conditions, Biochim Biophys Acta, 2013, 1827 (8–9), 986–1002. [DOI] [PubMed] [Google Scholar]
- 8. Capdevila D. A., Edmonds K. A., Giedroc D. P., Metallochaperones and metalloregulation in bacteria, Essays Biochem., 2017, 61, 177–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Homer M. J., Dean D. R., Roberts G. P., Characterization of the γ Protein and its involvement in the metallocluster assembly and maturation of dinitrogenase from Azotobacter vinelandii, J. Biol. Chem., 1995, 270 (42), 24745–24752. [DOI] [PubMed] [Google Scholar]
- 10. Rubio L. M., Rangaraj P., Homer M. J., Roberts G. P., Ludden P. W., Cloning and mutational analysis of the γ gene from Azotobacter vinelandii defines a new family of proteins capable of metallocluster binding and protein stabilization, J. Biol. Chem., 2002, 277 (16), 14299–14305. [DOI] [PubMed] [Google Scholar]
- 11. Lerner-Ellis J. P., Dobson C. M., Wai T., Watkins D., Tirone J. C., Leclerc D., Doré C., Lepage P., Gravel R. A., Rosenblatt D. S., Mutations in the MMAA gene in patients with the cblA disorder of vitamin B12 metabolism, Hum. Mutat., 2004, 24 (6), 509–516. [DOI] [PubMed] [Google Scholar]
- 12. Dobson C. M., Wai T., Leclerc D., Kadir H., Narang M., Lerner-Ellis J. P., Hudson T. J., Rosenblatt D. S., Gravel R. A., Identification of the gene responsible for the cblB complementation group of vitamin B12-dependent methylmalonic aciduria, Hum. Mol. Genet., 2002, 11 (26), 3361–3369. [DOI] [PubMed] [Google Scholar]
- 13. Yang X., Koohi-Moghadam M., Wang R., Chang Y.-Y., Woo P.C.Y., Li H., Sun H., Metallochaperone UreG serves as a new target for design of urease inhibitor: a novel strategy for development of antimicrobials, PLoS Biol., 2018, 16 (1), e2003887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fong Y. H., Wong H. C., Yuen M. H., Lau P. H., Chen Y. W., Wong K.-B., Structure of UreG/UreF/UreH complex reveals how urease accessory proteins facilitate maturation of Helicobacter pylori urease, PLoS Biol., 2013, 11 (10), e1001678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Phillips A. H., Hernandez J. A., Payá-Tormo L., Burén S., Cuevas-Zuviría B., Pacios L. F., Pelton J. G., Wemmer D. E., Rubio L. M., Environment and coordination of FeMo–co in the nitrogenase metallochaperone NafY, RSC Chem. Biol. 2021, 2 (5), 1462–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nonaka A., Yamamoto H., Kamiya N., Kotani H., Yamakawa H., Tsujimoto R., Fujita Y., Accessory proteins of the nitrogenase assembly, NifW, NifX/NafY, and NifZ, are essential for diazotrophic growth in the nonheterocystous cyanobacterium Leptolyngbya boryana, Front. Microbiol., 2019, 10, 495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Robinson N. J., Winge D. R., Copper Metallochaperones, Annu. Rev. Biochem., 2010, 79 (1), 537–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Leipe D. D., Wolf Y. I., Koonin E. V., Aravind L., Classification and evolution of P-loop GTPases and related ATPases, J. Mol. Biol., 2002, 317 (1), 41–72. [DOI] [PubMed] [Google Scholar]
- 19. Nim Y. S., Wong K.-B., The maturation pathway of nickel urease, Inorganics, 2019, 7 (7), 85. [Google Scholar]
- 20. Farrugia M. A., Macomber L., Hausinger R. P., Biosynthesis of the urease metallocenter, J. Biol. Chem., 2013, 288 (19), 13178–13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lacasse M. J., Zamble D. B., [NiFe]-hydrogenase maturation, Biochemistry, 2016, 55 (12), 1689–1701. [DOI] [PubMed] [Google Scholar]
- 22. Banerjee R., Gherasim C., Padovani D., The tinker, tailor, soldier in intracellular B12 trafficking, Curr. Opin. Chem. Biol., 2009, 13 (4), 484–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Banerjee R., B12 trafficking in mammals: a case for coenzyme escort service, ACS Chem. Biol. 2006, 1 (3), 149–159. [DOI] [PubMed] [Google Scholar]
- 24. Korotkova N., Lidstrom M. E., MeaB is a component of the methylmalonyl-CoA mutase complex required for protection of the enzyme from inactivation, J. Biol. Chem., 2004, 279 (14), 13652–13658. [DOI] [PubMed] [Google Scholar]
- 25. Jordan M. R., Wang J., Weiss A., Skaar E. P., Capdevila D. A., Giedroc D. P., Mechanistic insights into the metal-dependent activation of ZnII-dependent metallochaperones, Inorg. Chem., 2019, 58 (20), 13661–13672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sydor A. M., Jost M., Ryan K. S., Turo K. E., Douglas C. D., Drennan C. L., Zamble D. B., Metal binding properties of Escherichia coli YjiA, a member of the metal homeostasis-associated COG0523 family of GTPases, Biochemistry, 2013, 52 (10), 1788–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lonergan Z. R., Skaar E. P., Nutrient Zinc at the host–pathogen interface, Trends Biochem. Sci, 2019, 44 (12), 1041–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rodionov D. A., Vitreschak A. G., Mironov A. A., Gelfand M. S., Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes, J. Biol. Chem., 2003, 278 (42), 41148–41159. [DOI] [PubMed] [Google Scholar]
- 29. Osman D., Cooke A., Young T. R., Deery E., Robinson N. J., Warren M. J., The requirement for cobalt in vitamin B12: a paradigm for protein metalation, Biochim. Biophys. Acta., 2020, 118896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Crouzet J., Levy-Schil S., Cameron B., Cauchois L., Rigault S., Rouyez M. C., Blanche F., Debussche L., Thibaut D., Nucleotide sequence and genetic analysis of a 13.1-kilobase-pair pseudomonas denitrificans DNA fragment containing five cob genes and identification of structural genes encoding Cob (I) alamin adenosyltransferase, cobyric acid synthase, and bifunctional cobinamide kinase-cobinamide phosphate guanylyltransferase, J. Bacteriol., 1991, 173 (19), 6074–6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhou Z., Hashimoto Y., Cui T., Washizawa Y., Mino H., Kobayashi M., Unique biogenesis of high-molecular mass multimeric metalloenzyme nitrile hydratase: intermediates and a proposed mechanism for self-subunit swapping maturation, Biochemistry, 2010, 49 (44), 9638–9648. [DOI] [PubMed] [Google Scholar]
- 32. Sprang S. R., G PROTEIN MECHANISMS: insights from structural analysis, Annu. Rev. Biochem., 1997, 66 (1), 639–678. [DOI] [PubMed] [Google Scholar]
- 33. Hubbard P. A., Padovani D., Labunska T., Mahlstedt S. A., Banerjee R., Drennan C. L., Crystal structure and mutagenesis of the metallochaperone MeaB: insight into the causes of methylmalonic aciduria, J. Biol. Chem., 2007, 282 (43), 31308–31316. [DOI] [PubMed] [Google Scholar]
- 34. Lofgren M., Padovani D., Koutmos M., Banerjee R., A switch III motif relays signaling between a B12 enzyme and its G-protein chaperone, Nat. Chem. Biol., 2013, 9 (9), 535–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Froese D. S., Kochan G., Muniz J. R. C., Wu X., Gileadi C., Ugochukwu E., Krysztofinska E., Gravel R. A., Oppermann U., Yue W. W., Structures of the human GTPase MMAA and vitamin B12-dependent methylmalonyl-CoA mutase and insight into their complex formation, J. Biol. Chem., 2010, 285 (49), 38204–38213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jost M., Cracan V., Hubbard P. A., Banerjee R., Drennan C. L., Visualization of a radical B12 enzyme with its G-protein chaperone, Proc. Natl. Acad. Sci. 2015, 112 (8), 2419–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gasper R., Scrima A., Wittinghofer A., Structural insights into HypB, a GTP-binding protein that regulates metal binding, J. Biol. Chem., 2006, 281 (37), 27492–27502. [DOI] [PubMed] [Google Scholar]
- 38. Sydor A. M., Lebrette H., Ariyakumaran R., Cavazza C., Zamble D. B., Relationship between Ni(II) and Zn(II) coordination and nucleotide binding by the Helicobacter pylori [NiFe]-hydrogenase and urease maturation factor HypB, J. Biol. Chem., 2014, 289 (7), 3828–3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yuen M. H., Fong Y. H., Nim Y. S., Lau P. H., Wong K.-B., Structural insights into how GTP-dependent conformational changes in a metallochaperone UreG facilitate urease maturation, Proc. Natl. Acad. Sci., 2017, 114 (51), E10890–E10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Calixto A. R., Moreira C., Pabis A., Kötting C., Gerwert K., Rudack T., Kamerlin S. C. L., GTP Hydrolysis without an active site base: a unifying mechanism for Ras and related GTPases, J. Am. Chem. Soc., 2019, 141 (27), 10684–10701. [DOI] [PubMed] [Google Scholar]
- 41. Carvalho A. T. P., Szeler K., Vavitsas K., Åqvist J., Kamerlin S. C. L., Modeling the mechanisms of biological GTP hydrolysis, Arch. Biochem. Biophys., 2015, 582, 80–90. [DOI] [PubMed] [Google Scholar]
- 42. Wittinghofer A., Vetter I. R., Structure-function relationships of the G domain, a canonical switch motif, Annu. Rev. Biochem., 2011, 80 (1), 943–971. [DOI] [PubMed] [Google Scholar]
- 43. Li G., Zhang X. C., GTP hydrolysis mechanism of Ras-like GTPases, J. Mol. Biol., 2004, 340 (5), 921–932. [DOI] [PubMed] [Google Scholar]
- 44. Alfano M., Cavazza C., Structure, function, and biosynthesis of nickel-dependent enzymes, Protein Sci. 2020, 29 (5), 1071–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Douglas C. D., Ngu T. T., Kaluarachchi H., Zamble D. B., Metal transfer within the Escherichia coli HypB–HypA complex of hydrogenase accessory proteins, Biochemistry, 2013, 52 (35), 6030–6039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Takahashi-Iñiguez T., González-Noriega A., Michalak C., Flores M. E., Human MMAA induces the release of inactive cofactor and restores methylmalonyl-CoA mutase activity through their complex formation, Biochimie, 2017, 142, 191–196. [DOI] [PubMed] [Google Scholar]
- 47. Padovani D., Banerjee R., A G-protein editor gates coenzyme B12 loading and is corrupted in methylmalonic aciduria, Proc. Natl. Acad. Sci., 2009, 106 (51), 21567–21572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Padovani D., Banerjee R., Assembly and protection of the radical enzyme, methylmalonyl-CoA mutase, by its chaperone, Biochemistry, 2006, 45 (30), 9300–9306. [DOI] [PubMed] [Google Scholar]
- 49. Padovani D., Labunska T., Banerjee R., Energetics of interaction between the G-protein chaperone, MeaB, and B12-dependent methylmalonyl-CoA mutase, J. Biol. Chem., 2006, 281 (26), 17838–17844. [DOI] [PubMed] [Google Scholar]
- 50. Edmonds K. A., Jordan M. R., Giedroc D. P., COG0523 proteins: a functionally diverse family of transition metal-regulated G3E P-loop GTP hydrolases from bacteria to man, Metallomics, 2021, 13 (8), mfab046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lankathilaka K. P. W., Bennett B., Holz R. C., The Fe-type nitrile hydratase from Rhodococcus equi TG328-2 forms an alpha-activator protein complex, J. Biol. Inorg. Chem., 2020, 25 (6), 903–911. [DOI] [PubMed] [Google Scholar]
- 52. Gumataotao N., Lankathilaka K. P. W., Bennett B., Holz R. C., The iron-type nitrile hydratase activator protein is a GTPase, Biochem. J., 2017, 474 (2), 247–258. [DOI] [PubMed] [Google Scholar]
- 53. Hashimoto Y., Ube Y., Doi S., Kumano T., Kobayashi M., Metal chaperone, NhpC, involved in the metallocenter biosynthesis of nitrile hydratase, J. Gen. Appl. Microbiol., 2021, 67 (1), 24–32. [DOI] [PubMed] [Google Scholar]
- 54. Keeran N. S., Usha B., Ganesan G., P-type ATPases and their role in metal homeostasis in plants, In Metal and Nutrient Transporters in Abiotic Stress, 2021, Elsevier, pp. 33–54. [Google Scholar]
- 55. Dutkiewicz R., Nowak M., Craig E. A., Marszalek J., Fe–S cluster Hsp70 chaperones: the ATPase cycle and protein interactions, Methods Enzymol., 2017, 595, 161–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jeon W. B., Cheng J., Ludden P. W., Purification and characterization of membrane-associated CooC protein and its functional role in the insertion of nickel into carbon monoxide dehydrogenase from Rhodospirillum rubrum, J. Biol. Chem., 2001, 276 (42), 38602–38609. [DOI] [PubMed] [Google Scholar]
- 57. Jeoung J.-H., Goetzl S., Hennig S. E., Fesseler J., Wörmann C., Dendra J., Dobbek H., The extended reductive acetyl-CoA pathway: ATPases in metal cluster maturation and reductive activation, Biol. Chem., 2014, 395 (5), 545–558. [DOI] [PubMed] [Google Scholar]
- 58. Wittenborn E. C., Cohen S. E., Merrouch M., Léger C., Fourmond V., Dementin S., Drennan C. L., Structural insight into metallocofactor maturation in carbon monoxide dehydrogenase, J. Biol. Chem., 2019, 294 (35), 13017–13026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Silberg J. J., Tapley T. L., Hoff K. G., Vickery L. E., Regulation of the HscA ATPase reaction cycle by the co-chaperone HscB and the iron-sulfur cluster assembly protein IscU, J. Biol. Chem., 2004, 279 (52), 53924–53931. [DOI] [PubMed] [Google Scholar]
- 60. Jeoung J.-H., Giese T., Grünwald M., Dobbek H., Crystal structure of the ATP-dependent maturation factor of Ni,Fe-containing carbon monoxide dehydrogenases, J. Mol. Biol., 2010, 396 (4), 1165–1179. [DOI] [PubMed] [Google Scholar]
- 61. Kim J. H., Alderson T. R., Frederick R. O., Markley J. L., Nucleotide-dependent interactions within a specialized Hsp70/Hsp40 complex involved in Fe–S cluster biogenesis, J. Am. Chem. Soc., 2014, 136 (33), 11586–11589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Maio N., Jain A., Rouault T. A., Mammalian iron–sulfur cluster biogenesis: recent insights into the roles of frataxin, acyl carrier protein and ATPase-mediated transfer to recipient proteins, Curr. Opin. Chem. Biol., 2020, 55, 34–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Burén S., Jiménez-Vicente E., Echavarri-Erasun C., Rubio L. M., Biosynthesis of nitrogenase cofactors, Chem. Rev., 2020, 120 (12), 4921–4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Saini A., Mapolelo D. T., Chahal H. K., Johnson M. K., Outten F. W., SufD and SufC ATPase activity are required for iron acquisition during in vivo Fe-S cluster formation on SufB, Biochemistry, 2010, 49 (43), 9402–9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Layer G., Gaddam S. A., Ayala-Castro C. N., Ollagnier-de Choudens S., Lascoux D., Fontecave M., Outten F. W., SufE transfers sulfur from SufS to SufB for iron-sulfur cluster assembly, J. Biol. Chem., 2007, 282 (18), 13342–13350. [DOI] [PubMed] [Google Scholar]
- 66. Chahal H. K., Dai Y., Saini A., Ayala-Castro C., Outten F. W., The SufBCD Fe−S scaffold complex Interacts with SufA for Fe−S cluster Transfer, Biochemistry, 2009, 48 (44), 10644–10653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hirabayashi K., Yuda E., Tanaka N., Katayama S., wasaki K., Matsumoto T., Kurisu G., Outten F. W., Fukuyama K., Takahashi Y., Wada K., Functional dynamics revealed by the structure of the SufBCD complex, a novel ATP-binding cassette (ABC) protein that serves as a scaffold for iron-sulfur cluster biogenesis. J. Biol. Chem., 2015, 290 (50), 29717–29731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Camire E. J., Grossman J. D., Thole G. J., Fleischman N. M., Perlstein D. L., The yeast Nbp35-Cfd1 cytosolic iron-sulfur cluster scaffold is an ATPase, J. Biol. Chem., 2015, 290 (39), 23793–23802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Netz D. J. A., Pierik A. J., Stümpfig M., Mühlenhoff U., Lill R., The Cfd1–Nbp35 complex acts as a scaffold for iron-sulfur protein assembly in the yeast cytosol, Nat. Chem. Biol., 2007, 3 (5), 278–286. [DOI] [PubMed] [Google Scholar]
- 70. Lill R., Dutkiewicz R., Freibert S. A., Heidenreich T., Mascarenhas J., Netz D. J., Paul V. D., Pierik A. J., Richter N., Stümpfig M., The role of mitochondria and the CIA machinery in the maturation of cytosolic and nuclear iron–sulfur proteins, Eur. J. Cell Biol., 2015, 94 (7–9), 280–291. [DOI] [PubMed] [Google Scholar]
- 71. Molé C. N., Dave K., Perlstein D. L., Methods to unravel the roles of ATPases in Fe—S cluster biosynthesis, In Dos Santos P. C. (ed.), Fe-S Proteins: Methods and Protocols, 2021, New York, NY: Springer US, pp. 155–171. [DOI] [PubMed] [Google Scholar]
- 72. Sasaki D., Watanabe S., Matsumi R., Shoji T., Yasukochi A., Tagashira K., Fukuda W., Kanai T., Atomi H., Imanaka T., Miki K., Identification and structure of a novel archaeal HypB for [NiFe] hydrogenase maturation, J. Mol. Biol., 2013, 425 (10), 1627–1640. [DOI] [PubMed] [Google Scholar]
- 73. Watanabe S., Kawashima T., Nishitani Y., Kanai T., Wada T., Inaba K., Atomi H., Imanaka T., Miki K., Structural basis of a Ni acquisition cycle for [NiFe] hydrogenase by Ni-metallochaperone HypA and its enhancer, Proc. Natl. Acad. Sci., 2015, 112 (25), 7701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sestok A. E., Linkous R. O., Smith A. T., Toward a mechanistic understanding of Feo-mediated ferrous iron uptake, Metallomics, 2018, 10 (7), 887–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lau C. K. Y., Krewulak K. D., Vogel H. J., Bacterial ferrous iron transport: the Feo system, FEMS Microbiol. Rev., 2015, 40 (2), 273–298. [DOI] [PubMed] [Google Scholar]
- 76. Ash M.-R., Maher M. J., Guss J. M., Jormakka M., The initiation of GTP hydrolysis by the G-domain of FeoB: insights from a transition-state complex structure, PLoS One, 2011, 6 (8), e23355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Guilfoyle A. P., Deshpande C. N., Vincent K., Pedroso M. M., Schenk G., Maher M. J., Jormakka M., Structural and functional analysis of a FeoB A143S G5 loop mutant explains the accelerated GDP release rate, FEBS J., 2014, 281 (9), 2254–2265. [DOI] [PubMed] [Google Scholar]
- 78. Shin M., Mey A. R., Payne S. M., Vibrio cholerae FeoB contains a dual nucleotide-specific NTPase domain essential for ferrous iron uptake, Proc. Natl. Acad. Sci., 2019, 116 (10), 4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Gómez-Garzón C., Payne S. M., Vibrio cholerae FeoB hydrolyzes ATP and GTP in vitro in the absence of stimulatory factors†, Metallomics 2020, 12 (12), 2065–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Shin M., Park J., Jin Y., Kim I. J., Payne S. M., Kim K. H., Biochemical characterization of bacterial FeoBs: a perspective on nucleotide specificity, Arch. Biochem. Biophys., 2020, 685, 108350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Velayudhan J., Hughes N. J., McColm A. A., Bagshaw J., Clayton C. L., Andrews S. C., Kelly D. J., Iron acquisition and virulence in Helicobacter pylori: a major role for FeoB, a high-affinity ferrous iron transporter, Mol. Microbiol., 2000, 37 (2), 274–286. [DOI] [PubMed] [Google Scholar]
- 82. Marlovits T. C., Haase W., Herrmann C., Aller S. G., Unger V. M., The membrane protein FeoB contains an intramolecular G protein essential for Fe(II) uptake in bacteria, Proc. Natl. Acad. Sci., 2002, 99 (25), 16243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Waldron K. J., Robinson N. J., How do bacterial cells ensure that metalloproteins get the correct metal?, Nat. Rev. Microbiol., 2009, 7 (1), 25–35. [DOI] [PubMed] [Google Scholar]
- 84. Foster A. W., Osman D., Robinson N. J., 2014, Metal preferences and metallation, J. Biol. Chem., 289 (41), 28095–28103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Rae T. D., Schmidt P. J., Pufahl R. A., Culotta V. C., O'Halloran T. V., Undetectable intracellular free copper: the requirement of a copper chaperone for superoxide dismutase, Science, 1999, 284 (5415), 805–808. [DOI] [PubMed] [Google Scholar]
- 86. Outten C. E., O'Halloran T. V., Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis, Science, 2001, 292 (5526), 2488–2492. [DOI] [PubMed] [Google Scholar]
- 87. Dann C. E., Wakeman C. A., Sieling C. L., Baker S. C., Irnov I., Winkler W. C., Structure and mechanism of a metal-sensing regulatory RNA, Cell, 2007, 130 (5), 878–892. [DOI] [PubMed] [Google Scholar]
- 88. Xiao Z., Wedd A. G., The challenges of determining metal–protein affinities, Nat. Prod. Rep., 2010, 27 (5), 768–789. [DOI] [PubMed] [Google Scholar]
- 89. Young T. R., Martini M. A., Foster A. W., Glasfeld A., Osman D., Morton R. J., Deery E., Warren M. J., Robinson N. J., Calculating metalation in cells reveals CobW acquires Co II for vitamin B12 biosynthesis while related proteins prefer Zn II, Nat. Commun., 2021, 12 (1), 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Osman D., Martini M. A., Foster A. W., Chen J., Scott A. J. P., Morton R. J., Steed J. W., Lurie-Luke E., Huggins T. G., Lawrence A. D., Deery E., Warren M. J., Chivers P. T., Robinson N. J., Bacterial sensors define intracellular free energies for correct enzyme metalation, Nat. Chem. Biol., 2019, 15 (3), 241–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Junop M. S., Obmolova G., Rausch K., Hsieh P., Yang W., Composite active site of an ABC ATPase: MutS uses ATP to verify mismatch recognition and authorize DNA repair, Mol. Cell, 2001, 7 (1), 1–12. [DOI] [PubMed] [Google Scholar]
- 92. Krishnan A., Burroughs A. M., Iyer L. M., Aravind L., Comprehensive classification of ABC ATPases and their functional radiation in nucleoprotein dynamics and biological conflict systems, Nucleic Acids Res., 2020, 48 (18), 10045–10075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Khorasani-Motlagh M., Noroozifar M., Kerman K., Zamble D. B., Complex formation between the Escherichia coli [NiFe]-hydrogenase nickel maturation factors, Biometals 2019, 32 (3), 521–532. [DOI] [PubMed] [Google Scholar]
- 94. Takahashi-Íñiguez T., García-Arellano H., Trujillo-Roldán M. A., Flores M. E., Protection and reactivation of human methylmalonyl-CoA mutase by MMAA protein, Biochem. Biophys. Res. Commun., 2011, 404 (1), 443–447. [DOI] [PubMed] [Google Scholar]
- 95. Wu M., Lander G. C., How low can we go? Structure determination of small biological complexes using single-particle cryo-EM, Curr. Opin. Struct. Biol., 2020, 64, 9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Dempsey-Nunez L., Illson M. L., Kent J., Huang Q., Brebner A., Watkins D., Gilfix B. M., Wittwer C. T., Rosenblatt D. S., High resolution melting analysis of the MMAA gene in patients with cblA and in those with undiagnosed methylmalonic aciduria, Mol. Genet. Metab., 2012, 107 (3), 363–367. [DOI] [PubMed] [Google Scholar]
- 97. Beilschmidt L. K., Puccio H. M., Mammalian Fe–S cluster biogenesis and its implication in disease, Biochimie, 2014, 100, 48–60. [DOI] [PubMed] [Google Scholar]
- 98. Mandal S. K., Nayak S. G., Kanaujia S. P., Identification and characterization of metal uptake ABC transporters in Mycobacterium tuberculosis unveil their ligand specificity, Int. J. Biol. Macromol., 2021, 185, 324–337. [DOI] [PubMed] [Google Scholar]
- 99. Chan K. H., Li T., Wong C. O., Wong K. B., Structural basis for GTP-dependent dimerization of hydrogenase maturation factor HypB, PLoS One, 2012, 7 (1), e30547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Cupp-Vickery J. R., Peterson J. C., Ta D. T., Vickery L. E., Crystal structure of the molecular chaperone HscA cubstrate binding domain complexed with the IscU recognition peptide ELPPVKIHC, J. Mol. Biol., 2004, 342 (4), 1265–1278. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No new data were generated or analyzed in support of this research.