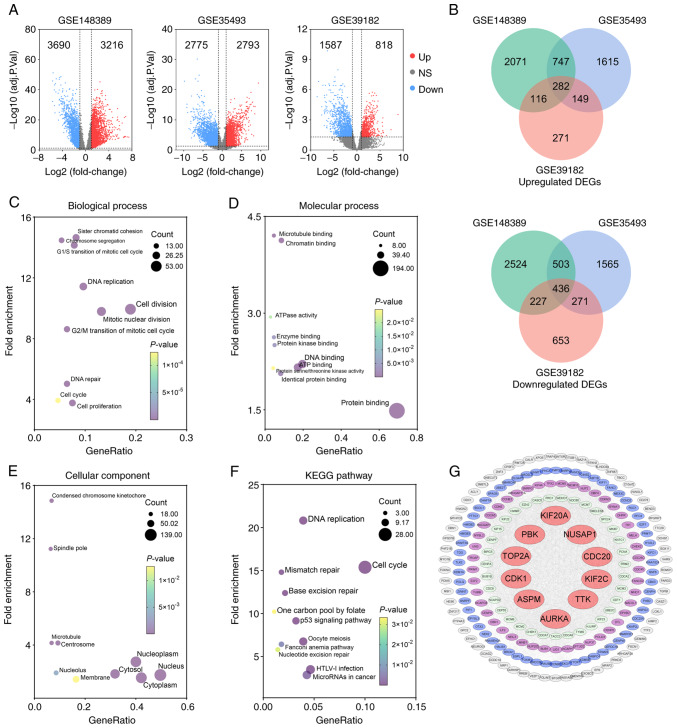
Figure 2.
Identification of upregulated hub genes in MB following enrichment analysis and protein-protein interaction network construction. (A) Volcano plots illustrating upregulated DEGs (red dots) and downregulated DEGs (blue dots) identified in the three datasets with the criteria of adjusted P<0.05 and |log2FC| ≥1 (11–13). (B) Venn diagram of overlapping upregulated or downregulated DEGs among three datasets. (C-E) GO analysis of upregulated DEGs presents the top 10 significant terms of GO analysis in (C) biological process, (D) molecular function, and (E) celluar component. (F) Significant KEGG pathway enrichment terms. (G) The PPI network was constructed with upregulated genes. Red circular nodes represent the top 10 hub genes. MB, medulloblastoma; DEGs, differentially upregulated genes; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; PPI, protein-protein interaction; PBK, PDZ binding kinase; CDC20, cell division cycle 20; KIF2C, kinesin family member 2C; NUSAP1, nucleolar and spindle associated protein 1; TTK, TTK protein kinase; KIF20A, kinesin family member 20A; TOP2A, DNA topoisomerase II alpha; CDK1, cyclin dedendent kinase 1; ASPM, assembly factor for spindle microtubules; AURKA, Aurora kinase A.