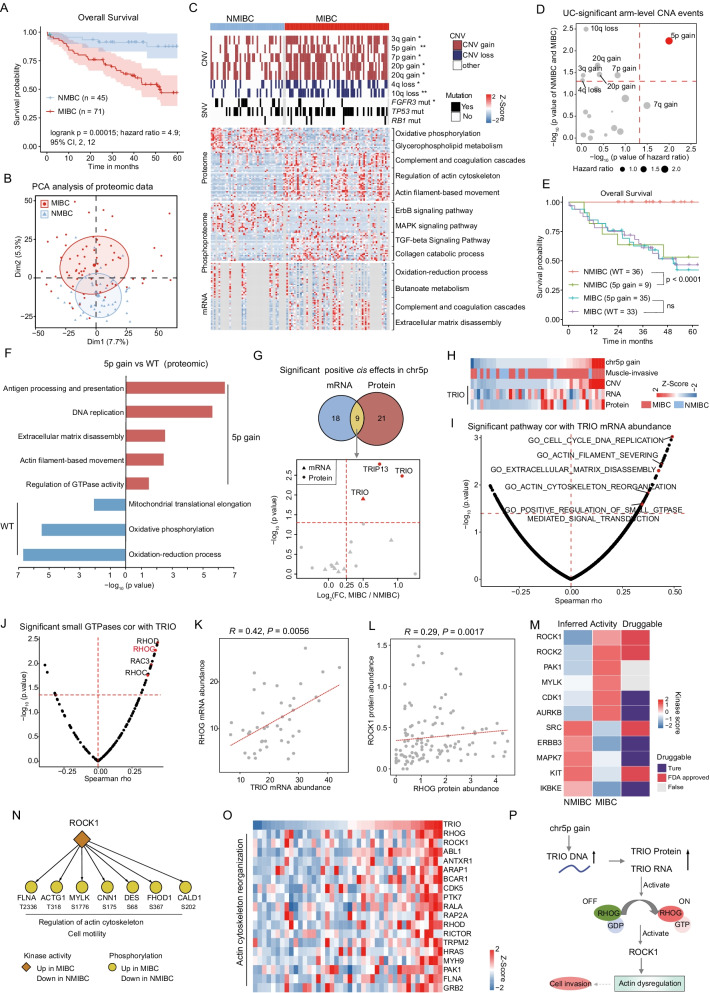
Fig. 4.
Proteogenomic profiles distinguished NMIBC from MIBC. A Overall survival analysis of NMIBC versus MIBC patients (p value from log-rank test). 95% confidence interval was also presented. B PCA analysis of proteomic data (5683 proteins) between MIBC and NMIBC. Red dots: MIBC; blue dots: NMIBC. C Differentially variational genomic events (top panel) and differentially expressed genes, proteins and phosphoproteins in MIBC and NMIBC and their associated biological pathways (bottom panel). Fisher’s exact test was used for arm-level can events and the status of genes mutation. The Wilcoxon rank-sum test was used for differential expression analysis. D Significantly different arm-level CNA events in MIBC and NMIBC and their association with prognosis. E Survival analysis of NMIBC and MIBC patients with chromosome 5p gain versus WT (p value from log-rank test). F Pathways enriched in differentially expressed proteins between 5p gain and WT. G Overlap of genes with significant positive cis-effect genes on 5p based on RNA-Seq or proteomic data (top panel) and log2-fold change between NMIBC and MIBC were shown for the nine overlapping genes (bottom panel). The dots represent proteins; the triangles represent mRNA. H Heatmap of copy number gain of 5p and the mRNA/protein abundance of TRIO. I Volcano plot showing the correlation between enriched Gene Ontology biological processes and TRIO mRNA abundance. J Volcano plot showing the correlation between small GTPases and TRIO based on mRNA level. The one highlighted in red is reportedly activated by TRIO. K Correlation of TRIO mRNA abundance with RHOG mRNA abundance. L Correlation of RHOG protein abundance with ROCK1 protein abundance. M Evaluation of kinase activities in tumors of NMIBC and MIBC via KSEA. Drug targets were based on the Drug Gene Interaction Database (http://www.dgidb.org/). N Diagram illustrating differences between NMIBC and MIBC tumors in terms of phosphorylation abundance and kinase activity for ROCK1. O Heatmap of the mRNA abundance of actin cytoskeleton reorganization related genes. P A brief model depicting the functional impact of chromosome 5p gain. The p values in I–L were calculated by Spearman's correlation test