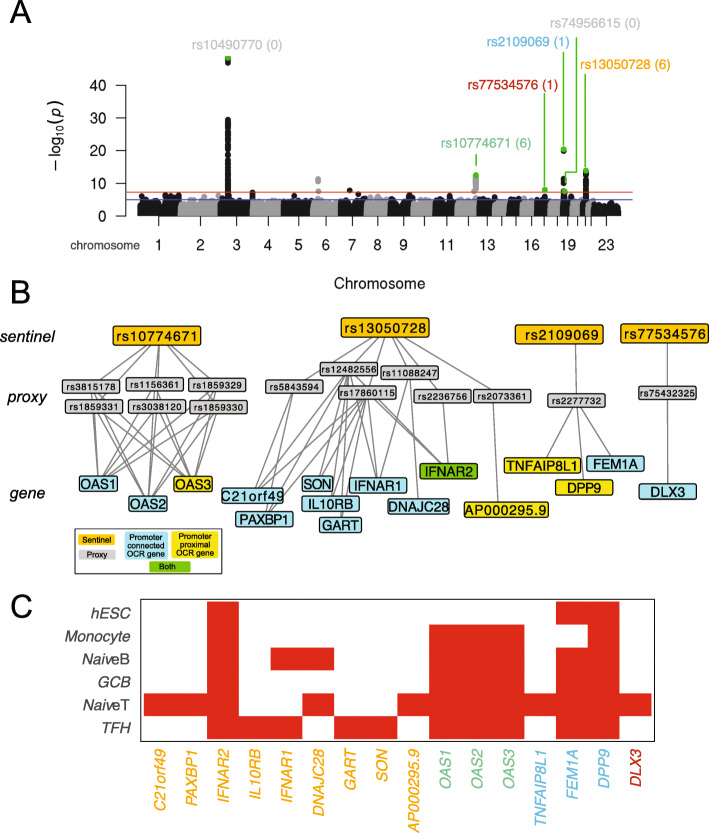
Fig. 2.
Chromosome capture-based variant-to-gene mapping identifies candidate effector genes at COVID-19 GWAS loci. A Manhattan plot generated using the summary statistics from the COVID-19 severity GWAS. Genome-wide significant signals are shown together with the number of accessible gene-annotated proxies associated. B Depiction of the statistical sentinel-proxy SNP linkages and the PCC-derived physical gene-proxy connections identified in this study. Genes in yellow were implicated by an accessible proxy in the promoter regions, genes in blue were implicated through chromatin-based contact between the promoter region and a distal accessible proxy, and green indicates implication by both promoter and distal proxies. C Heatmap depicting genes implicated by variant-to-gene mapping in each cell type in red. Color of each gene corresponds to the signal shown in A