Fig. 4.
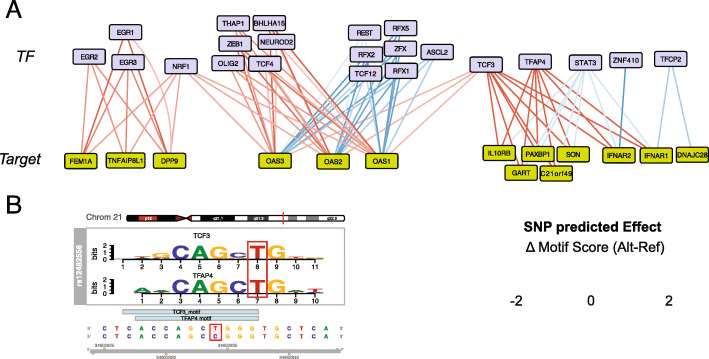
In silico prediction of transcription factor binding site disruption by accessible COVID-19 associated proxies. A Transcription factors (blue) with binding motifs likely to be disrupted by accessible COVID-19 SNPs and their connected target genes (green). The predicted effect of the SNP on TF binding is indicated in red for decreased affinity and in blue for increased affinity. COVID-19 risk-associated sequence variation at an element connected to IFNAR2 is predicted to increase binding of ZNF410, a zinc finger protein involved in repression of fetal hemoglobin in erythroid cells. Risk variants also increased the predicted affinity of STAT3 for elements at six implicated genes including PAXBP1. Risk variants at PAXBP1 and five other implicated genes were also predicted to reduce binding of the MYC-induced AP4 (TFAP4) oncoprotein and E2A (TCF3), a central transcription factor in lymphocyte development and malignancy. COVID-19 disease variants connected to OAS1 and OAS3 were predicted to affect the binding of 15 distinct transcription factors, including the E proteins TCF3, TCF4, TCF12, and NEUROD2, the RFX family of transcription factors involved in expression of a variety of immune factors including the MHC class II genes, and the plasmablast and TFH factors MIST1 (BHLHA15) and ASCL2. B An example of the predicted impact of the COVID-19 risk allele of rs12482556 (red) on binding of TCF3 and TFAP4