Myelin genes are amongst the most dysregulated genes in the aged brain: Just over half of the weight of an entire adult human brain is attributed to myelin, which wraps around neuronal axons and is essential for superfast axonal conduction and neuronal integrity. In the central nervous system, it is the function of specialized cells called oligodendrocytes (OLs) to make myelin, which is made up of lipids and proteins. OLs are generated throughout life by a significant population of oligodendrocyte progenitor cells (OPCs) that are responsible for the lifelong generation of OLs and myelin, essential for learning, as well as repair following pathological insults (i.e. in demyelinating diseases that include multiple sclerosis) (Simons and Nave, 2015; Philips and Rothstein, 2017). Changes in myelin content in the human brain over the lifespan of individuals have been well documented, as well as evidence of myelin loss in rodent models using classical histological approaches (Bartzokis et al., 2012; Soreq et al., 2017). During the normal course of aging, degenerative alterations in myelin (myelin thinning, formation of myelin balloons, loss of myelinated tracts) have been shown to precede overt neuronal loss and ultimately lead to negative clinical outcomes, which manifest as the dramatic decline in the speed and efficiency of neuronal networks [reviewed in Rivera et al. (2021a)]. However, a gap in our knowledge was the precise changes in OL and myelin genes at the transcriptome level, which can inform the genetic programs for targeting rejuvenation (Neumann et al., 2019; Rivera et al., 2021b).
In our recent study, we sought to address these issues by performing high-throughput transcriptomic profiling of global gene changes in the aged mouse cerebrum (Rivera et al., 2021b). In the first instance, the clearest cut genes belonged to those involved in myelination, which prompted further investigations to tease out the genetic programs in individual stages of OL lineage cells via a meta-analysis of publicly available datasets generated by the Barres lab (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE52564). These experiments revealed that genes expressed by OPCs and the major OL myelin genes are dramatically altered in aging. These include, but are not limited to, Mog, Plp1, Cnp, and Ugt8a, along with lesser-known myelin-specific genes, for example, Cldn11 and Gsn. Notably, as well as the assembly and synthesis of myelin being markedly perturbed as the brain ages, there was a near loss of transcriptional networks implicated in OPC turnover and lineage progression. In the adult brain through to aging, a small (2–5%) cohort of cells in the brain are OPCs, enabling myelin remodeling, which is most efficient in younger mice (Figure 1A and B) (Rivera et al., 2021b). Our study identified the key oligodendroglial gene networks that are most disrupted in aging and presented new targets for promoting myelin repair, as discussed further below.
Figure 1.
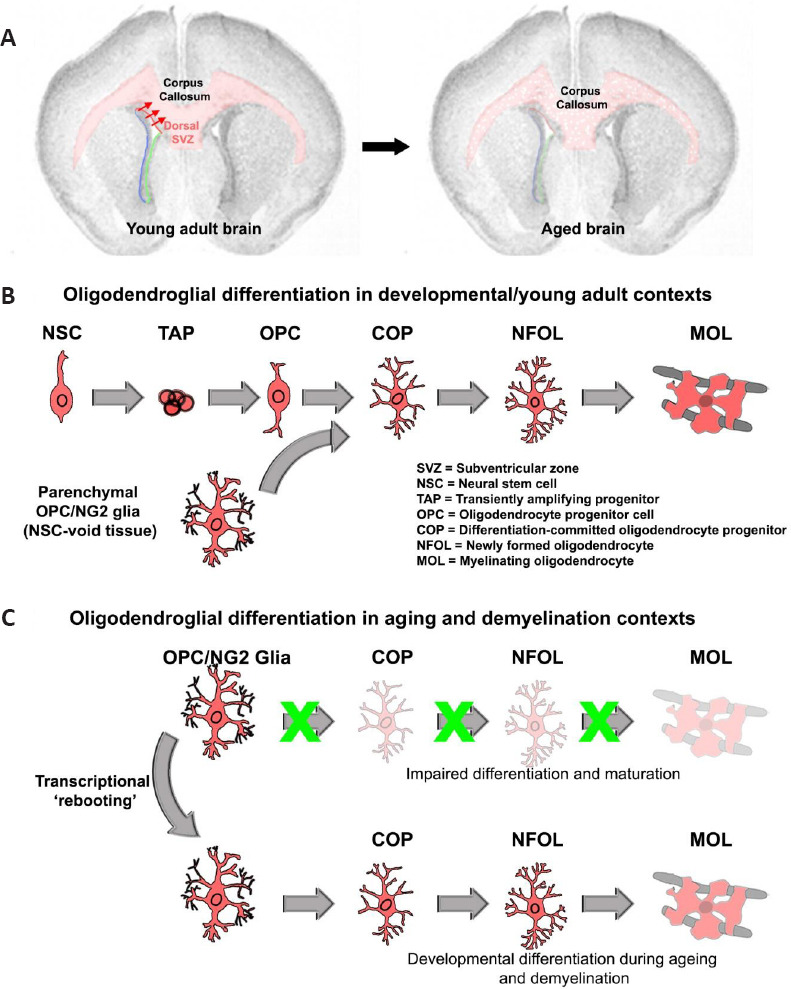
Impaired OL lineage differentiation during aging leads to loss of compact myelin.
(A) Coronal brain sections illustrating the effects of aging on myelin content, highlighting the major white matter region of the brain the corpus callosum exhibiting marked loss in myelin (in light red). In the young adult brain, the dorsal SVZ is an additional reservoir for oligodendrogenesis which is most active during developmental periods. (B) Overview of the major steps of differentiation of the OL lineage. In the young adult brain through to adulthood, parenchymal OPCs are the main pool responsible for myelin remodeling, although to a lesser degree, NSCs can also contribute to the generation of adult-born MOLs. (C) OPCs during aging (and following a demyelinating insult in aged mice) are inefficient in their capacity to differentiate along the OL lineage. Pharmacogenomically activating transcriptional networks associated with younger OPCs during aging, promotes lineage progression, leading to the appearance of MOLs and subsequent remyelination/myelin remodeling.
A key finding from our study was that the most downregulated gene during brain aging was the G-protein coupled Receptor 17 (Gpr17), which is expressed by a pool of late-stage OPCs that are committed to OL generation and are referred to as committed-OPCs (COPs). To pursue this further, we performed genetic fate-mapping of OPCs and COPs and their progeny in 3- versus 18-month old mice, using Pdgfra:::CreERT2 and Gpr17:::CreERT2 transgenic mice (Rivera et al., 2021b). The results revealed a striking loss of COPs and their generation of OLs in aged brains, whilst the overall density of OPCs remained relatively unchanged. These findings highlighted that aging dramatically perturbs a specific subset of OPCs, namely COPs, which are the prerequisite stage for the generation of myelinating OLs (Figure 1B and C). Our unbiased approach of resolving the age-related transcriptional hallmarks in OL lineage cells enabled the identification of new ways to ‘reboot’ transcriptional networks’ in aged OPCs and revert them back to their younger counterparts to rejuvenate myelination in the aging brain. In addition, although the genes associated with neuron subclasses, astrocytes, and microglia were also dysregulated as described by others (Ximerakis et al., 2019), their age-related transcriptional changes were lesser compared to oligodendroglial genes which was the focus of our study.
Systems biology for correcting impaired transcriptional networks associated with aged OPCs: The decline in OL lineage progression during aging is now known to be attributed to dysregulated transcriptional networks caused by lack of trophic support in the brain (Azim et al., 2018; Ximerakis et al., 2019). This raised the hypothesis that perturbed transcriptional networks in aged OPCs could be targeted by systems biology approaches to identify therapeutic agents for promoting transcriptional networks associated with younger OPCs. One such interdisciplinary approach is based on the accumulation of genomics and chemical informatics data that have emerged as novel strategies for addressing pharmacologically disease-induced transcriptional alterations. For this, we adopted the Connectivity Map and Library of Integrated Network-based Cellular Signatures (Azim et al., 2017). This resource offers the potential for identifying agents already in clinical use as novel therapies for aging and remyelination, which are currently clinically unmet needs. A unique feature of this drug discovery strategy relies on the very transcriptomic nature of the analysis, which enables the identification of drug-specific gene networks with precise mechanisms of action (Azim et al., 2017).
Having identified the aging signatures of OL lineage cells and validated these using cre-loxP of OPC- and COP-derived OLs, our next aim was to rejuvenate stem-like features in aged OPCs. To achieve this, we interrogated gene signatures of young versus aged OPC into recently optimized Connectivity Map and Library of Integrated Network-based Cellular Signatures databases for obtaining small molecules for promoting OPC stemness (Rivera et al., 2021c). In addition, the STITCH drug-protein interaction database (http://stitch.embl.de) was used to obtain any possible common small molecules for testing in vivo. An important finding was that small molecules that specifically stimulate OPCs to generate COPs were most distinct, compared to agents that promote differentiation of COPs into myelinating OLs (Figure 1B and C). Amongst the highest-ranked small molecules predicted to drive transcriptional phenotypes of young OPC, the small molecule LY294002 stood out because it is best known as a modulator of the PI3K/Akt/mTOR/PTEN signaling axis, which is known to be essential for OPC survival and differentiation (Rivera et al., 2021c). The next phase of our study was to re-examine the potential targets of this molecule and test its efficacy in a demyelination model.
Pharmacogenomic-targeting of developmentally regulated processes during myelination and remyelination in young adult mice: Revisiting LY294002 with newer chemical library databases revealed further protein targets aside from those classically defined and suggested that LY294002 targets a host of intracellular proteins in a dose-specific manner (for example: https://lincs.hms.harvard.edu/db/sm/10510-101-1/). The lower dosing of LY294002 was found in our recent study to be beneficial for OL differentiation when administered into the lateral ventricle. It should also be noted that at lower micromolar ranges, LY294002 induces entirely contrasting effects on OLs compared to more generally used higher doses of 20–30 μM that are detrimental to OLs (Rivera et al., 2021c). Presently, we have provided evidence that the bromodomain-containing proteins (Brd2-4) are novel unexplored targets of LY294002 (Rivera et al., 2021c).
Resetting transcriptional networks in aged OPCs toward their younger counterparts: Knowing from our most recent work that low doses of LY294002 readily promote OPC stemness and lineage progression in vivo in younger mice (Rivera et al., 2021c), we hypothesized that this small molecule was a promising candidate for reverting aged OPCs to their developmental phenotypes (Figure 1D). For these experiments, myelin regeneration was tested in older mice (> 6 months of age), at which age the remyelinating capacity of OPCs declines steeply and remains diminished thereafter. A demyelinating lesion was induced using lysolecithin followed by treatment with LY294002, which significantly increased OPC density and their differentiation into OLs, with a subsequent improvement in remyelination, that otherwise failed in the aged brain (Figure 1D). Notably, our analyses also identified a number of other small molecules that target multiple signaling pathways predicted to rejuvenate OPC regenerative capacity, and combinatorial therapies will likely result in more efficient remyelination in the aging brain. For example, sequential treatment with small molecules that first target transcriptional networks to increase the density of OPCs and COPs, then agents that specifically stimulate their differentiation downstream in the OL lineage, and finally target networks that promote myelination in newly formed OLs (Rivera et al., 2021b). Further studies are required to test these hypotheses and to determine how different treatment regimens affect the pro-myelinating actions of other cells (microglia, astrocytes, neurons).
Future perspectives: In summary, our latest findings highlight that oligodendroglial genes are amongst the most dysregulated during the aging of the brain and there is severe loss of transcriptional networks that are essential for the successful transition of OPCs into myelinating OLs. We propose that our systems biology approach provides an efficient and unbiased method for identifying key transcriptional networks that can be targeted to promote rejuvenation of aged oligodendroglia, since no single gene is a ‘silver bullet’ for controlling OPC numbers and differentiation, and it will not be possible to achieve regulation of scores of genes in clinical settings. A key aspect that requires attention in the field is the relative importance of OL heterogeneity distinct central nervous system compartments in aging, in both health and disease states. Since demyelinating diseases such as multiple sclerosis affect multiple central nervous system regions, often commencing in the spinal cord and/or optic nerve, it will be important to determine whether strategies that work in the mouse cerebrum also promote repair in other regions and in human tissue, for example in the optic nerve and cerebellar tissue (Rivera et al., 2021c). Overall, our results demonstrate massive deficits in OL lineage cells in natural aging that impact significantly on remyelination and repair, which is highly relevant to neuropathology, including multiple sclerosis and Alzheimer’s disease.
We apologize to the experts in the field who have made important contributions for not discussing their work fully due to space limitations.
This work was supported by a PhD Studentship from The Anatomical Society (to ADR, AMB), and grants from the BBSRC (to AMB, ADR, Grant Number BB/M029379/1), MRC (to AMB, Grant Number MR/P025811/1), Multiple Sclerosis Society of the UK (to AMB; Award Reference: 40), MSCA Seal of Excellence @ UNIPD and NVIDIA Hardware Grant (to ADR), German Research Council (AZ/115/1-1; AZ/115/1-3), Swiss National Funds (to KA; P300PA_171224).
Footnotes
C-Editors: Zhao M, Zhao LJ, Qiu Y; T-Editor: Jia Y
References
- 1.Azim K, Akkermann R, Cantone M, Vera J, Jadasz JJ, Küry P. Transcriptional profiling of ligand expression in cell specific populations of the adult mouse forebrain that regulates neurogenesis. Front Neurosci. 2018;12:220. doi: 10.3389/fnins.2018.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Azim K, Angonin D, Marcy G, Pieropan F, Rivera A, Donega V, Cantu C, Williams G, Berninger B, Butt AM, Raineteau O. Pharmacogenomic identification of small molecules for lineage specific manipulation of subventricular zone germinal activity. PLoS Biol. 2017;15:e2000698. doi: 10.1371/journal.pbio.2000698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartzokis G, Lu PH, Heydari P, Couvrette A, Lee GJ, Kalashyan G, Freeman F, Grinstead JW, Villablanca P, Finn JP, Mintz J, Alger JR, Altshuler LL. Multimodal magnetic resonance imaging assessment of white matter aging trajectories over the lifespan of healthy individuals. Biol Psychiatry. 2012;72:1026–1034. doi: 10.1016/j.biopsych.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 4.Neumann B, Baror R, Zhao C, Segel M, Dietmann S, Rawji KS, Foerster S, McClain CR, Chalut K, van Wijngaarden P, Franklin RJM. Metformin restores CNS remyelination capacity by rejuvenating aged stem cells. Cell Stem Cell. 2019;25:473–485. doi: 10.1016/j.stem.2019.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Philips T, Rothstein JD. Oligodendroglia:metabolic supporters of neurons. J Clin Invest. 2017;127:3271–3280. doi: 10.1172/JCI90610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rivera AD, Chacon-De-La-Rocha I, Pieropan F, Papanikolau M, Azim K, Butt AM. Keeping the ageing brain wired:a role for purine signalling in regulating cellular metabolism in oligodendrocyte progenitors. Pflugers Arch. 2021a;473:775–783. doi: 10.1007/s00424-021-02544-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rivera AD, Pieropan F, Chacon-De-La-Rocha I, Lecca D, Abbracchio MP, Azim K, Butt AM. Functional genomic analyses highlight a shift in Gpr17-regulated cellular processes in oligodendrocyte progenitor cells and underlying myelin dysregulation in the aged mouse cerebrum. Aging Cell. 2021b;20:e13335. doi: 10.1111/acel.13335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rivera AD, Pieropan F, Williams G, Calzolari F, Butt AM, Azim K. Drug connectivity mapping and functional analysis reveal therapeutic small molecules that differentially modulate myelination. Biomed Pharmacother. 2021c;145:112436. doi: 10.1016/j.biopha.2021.112436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simons M, Nave KA. Oligodendrocytes:myelination and axonal support. Cold Spring Harb Perspect Biol. 2015;8:a020479. doi: 10.1101/cshperspect.a020479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soreq L, Rose J, Soreq E, Hardy J, Trabzuni D, Cookson MR, Smith C, Ryten M, Patani R, Ule J. Major shifts in glial regional identity are a transcriptional hallmark of human brain aging. Cell Rep. 2017;18:557–570. doi: 10.1016/j.celrep.2016.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ximerakis M, Lipnick SL, Innes BT, Simmons SK, Adiconis X, Dionne D, Mayweather BA, Nguyen L, Niziolek Z, Ozek C, Butty VL, Isserlin R, Buchanan SM, Levine SS, Regev A, Bader GD, Levin JZ, Rubin LL. Single-cell transcriptomic profiling of the aging mouse brain. Nat Neurosci. 2019;22:1696–1708. doi: 10.1038/s41593-019-0491-3. [DOI] [PubMed] [Google Scholar]


