Figure 3.
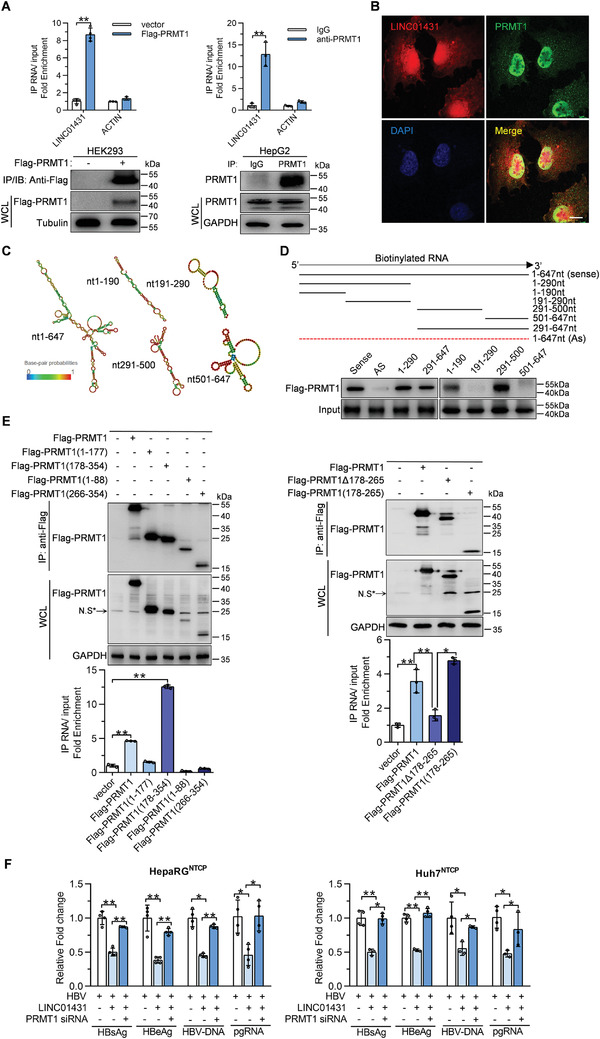
PRMT1 interacts with LINC01431 to mediate its anti‐HBV effect. A) RIP assay for LINC01431 in Flag‐PRMT1 overexpressed HEK293 and HepG2 cells using anti‐Flag (left) or anti‐PRMT1 (right). Normalized data were shown as relative fold enrichment to the control group. B) Immunofluorescence of LINC01431 and PRMT1 in Huh7 cells. Nuclei were stained with DAPI (blue). Scale bar, 20 µm. C) Predicted secondary structures of LINC01431 and its truncated mutants were presented. D) RNA‐pull down assays for the in vitro interaction between truncated LINC01431 mutants and PRMT1. Huh7 cells were transfected with Flag‐PRMT1, and the cell lysates were incubated with biotinylated LINC01431 transcripts, followed by immunoblotting with indicated antibodies. E) RIP assay for the in vivo interaction between LINC01431 and PRMT1. HepG2 cells were transfected with LINC01431 and plasmids expressing different Flag‐tagged‐PRMT1 truncates. RIP assay was performed using anti‐Flag antibody at day 3 post transfection. The level of PRMT1 and its mutants were detected by immunoblotting (upper). Normalized data were shown as relative fold enrichment to the control group (bottom). N.S* indicated the non‐specific band. F) Rescue assays were performed in HBV‐infected HepaRGNTCP cells (left) or HBV‐infected Huh7NTCP cells (right) after silencing PRMT1. The levels of HBV antigens, HBV‐DNA, and pgRNA were detected at 5 dpi by ELISA and RT‐qPCR, respectively. WCL indicated the whole cell lysis. GAPDH and Tubulin served as the loading control. For (A,B) and (D–F), representative of 3 independent experiments. Data information: data were presented as mean ± SD and normalized to the control group. One‐way ANOVA (F). Two‐tail unpaired Student's t‐tests; * p < 0.05; ** p < 0.01 (A,E).
