Fig. 1.
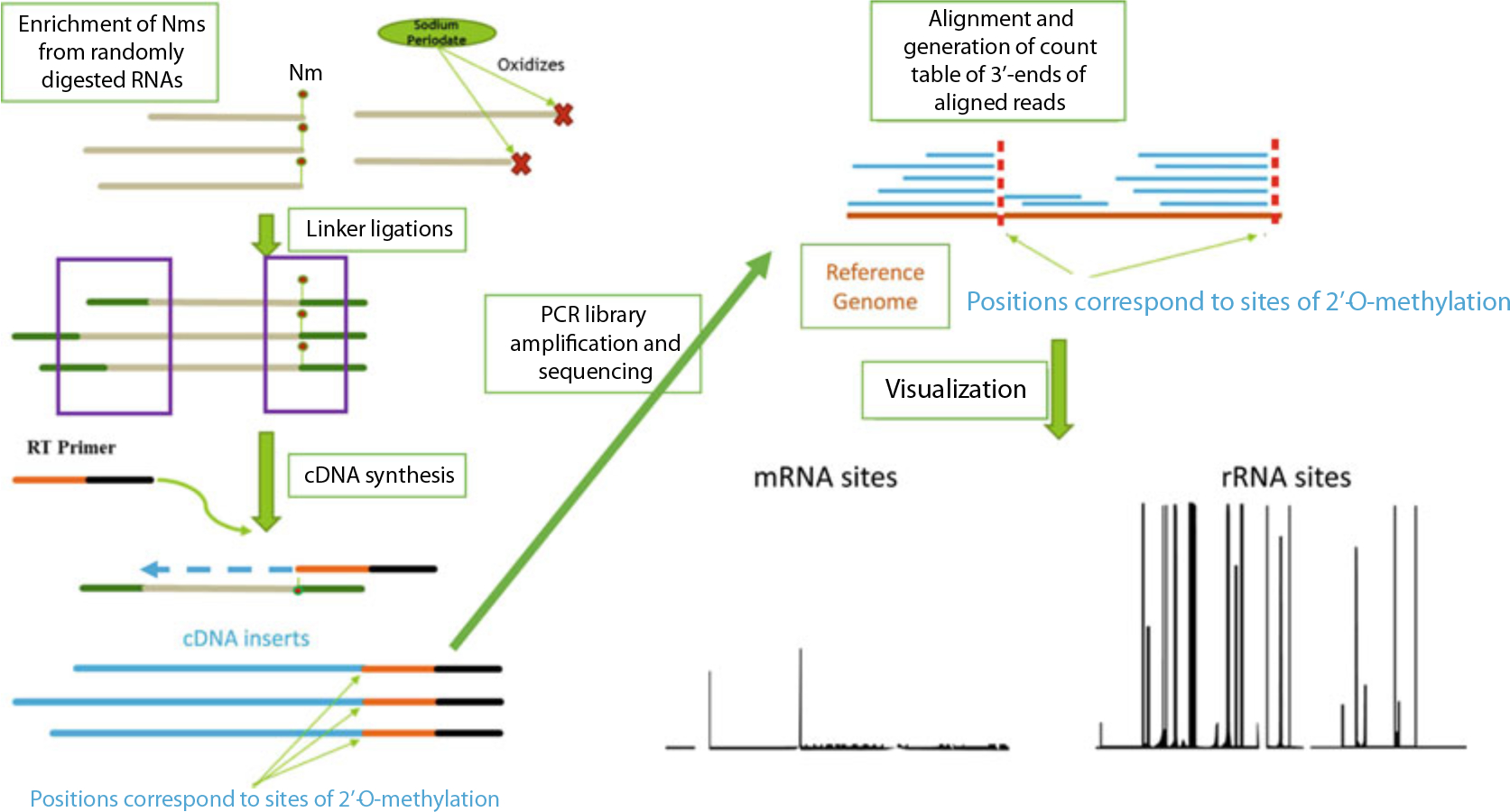
Concept of RibOxi-seq chemistry and computational pipeline. RNA input is randomly digested, followed by enrichment of Nm ends through oxidation of unmethylated 3′-ends by sodium periodate. Libraries containing only nonoxidized fragments are made through the ligation method. Libraries are sequenced on Illumina platforms of choice. Reads are then aligned to a reference genome to allow 3′-end pile-up counting. Positions with significant 3′-end counts corresponds to sites protected by Nm
