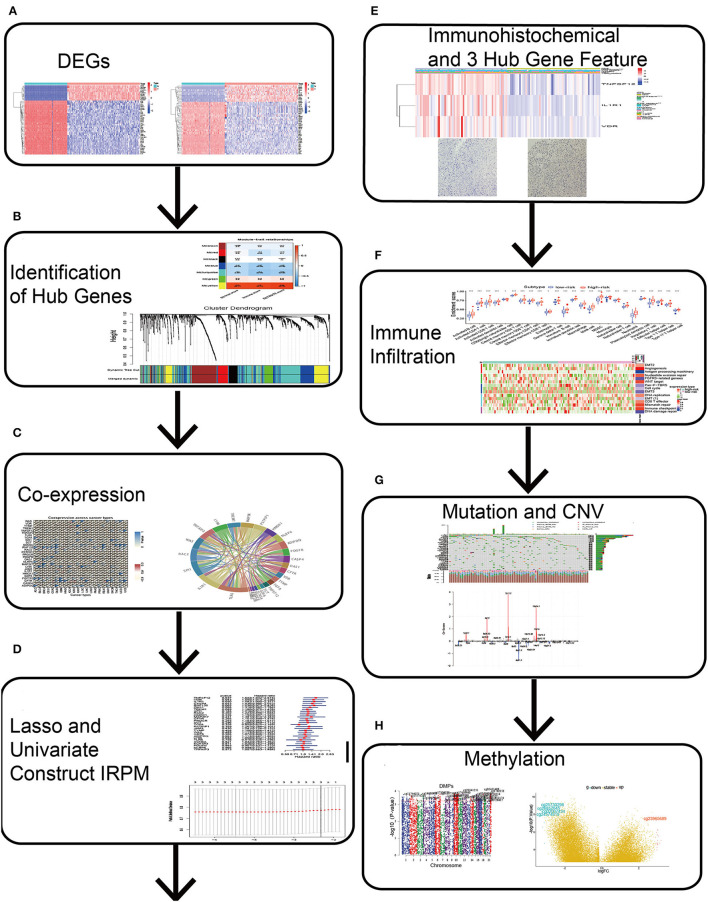
Figure 1.
Schematic diagram of this study. (A) Differential genes were obtained from GBM and normal brain tissue. (B) Hub genes were identified by Weighted Gene Co-Expression Network Analysis (WGCNA). (C) Clinicopathologic characteristics of 28 hub genes were analyzed. (D) Lasso and univariate regression were used to identify 3 key genes and the corresponding risk coefficients. (E) ICH and TIMER were used to analyse the clinicopathological characteristics of three key genes. (F) ssGSEA was used to analyse immune infiltration in both groups. (G) Maftools and Genomic Identification of Significant Targets in Cancer algorithm were used to analyse mutations and CNV in both groups. (H) ChAMP was used to analyse methylation in high and low-risk groups.