Fig. 5. Total biliary scRNA-seq reveals heightened inflammatory tone of biliary tissue in the absence of tuft cells.
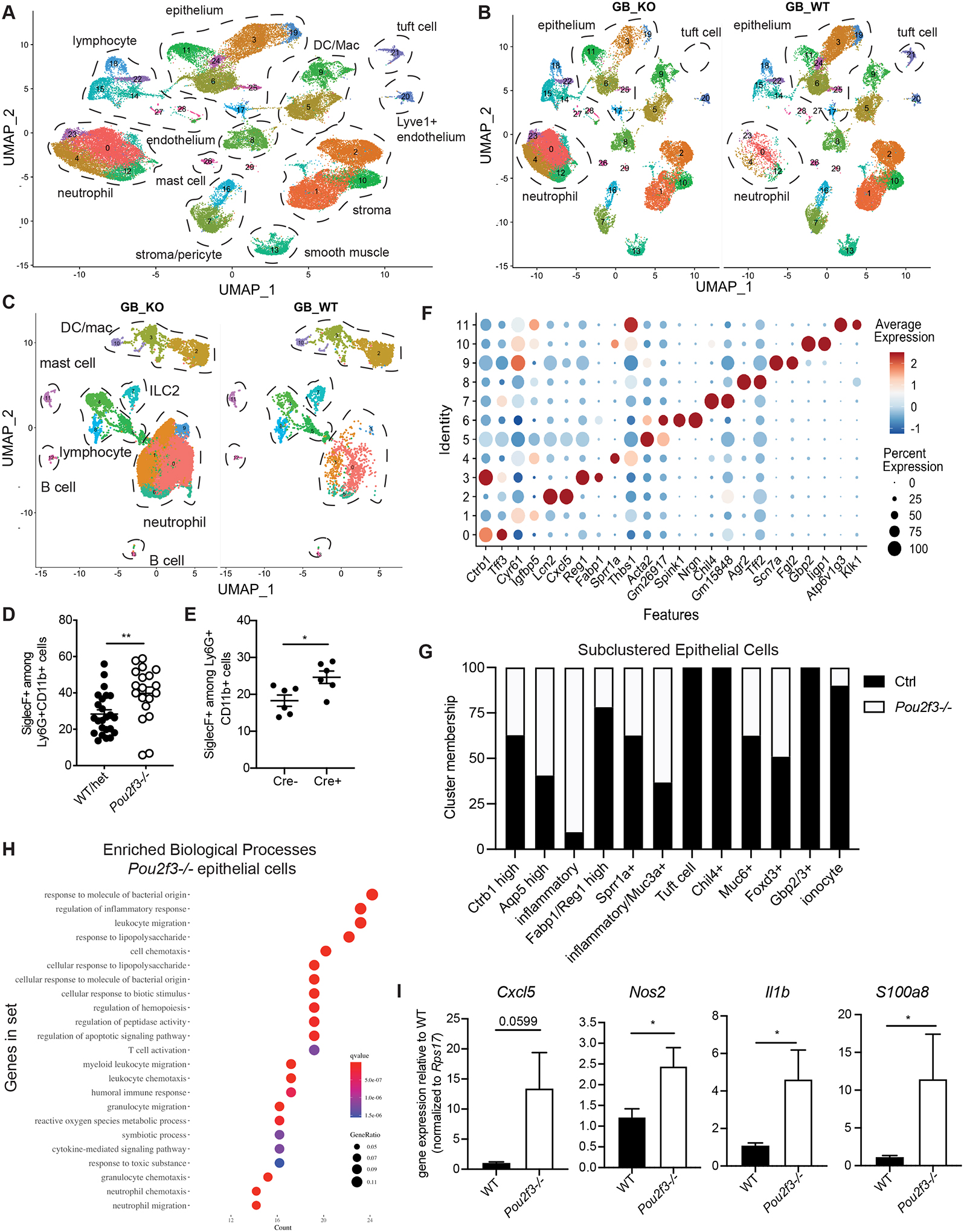
A-B) Live cells were sorted from Pou2f3−/− and littermate controls total GB/EHBD digests and subjected to single cell sequencing using the 10X platform. UMAP shows major cell types, determined by canonical gene expression. C) Subclustered CD45+ cells split by genotype: Pou2f3−/− (GB_KO), littermate controls (GB_WT) . D,E) Quantification of Siglec F+ neutrophils from littermate Pou2f3−/− and littermate controls (D) and 25cre/+;R26iDTR or 25+/+;R26iDTR/iDTR mice injected with diphtheria toxin at 4 wks of age and analyzed 4–6 wks later (E) as determined by flow cytometry on total GB/EHBD digests. F) Dotplot of top two most differentially expressed genes (DEGs) after subclustering of biliary epithelial cells. G) Epithelial cell cluster membership after normalizing to the total number of epithelial cells sequenced per genotype. H) Gene ontology biological processes enrichment for genes upregulated in epithelial cells from Pou2f3−/− mice compared to controls. I) Total GB/EHBD from non-littermate Pou2f3−/− (n=17) and WT (n=15) mice was analyzed by qPCR for the indicated target genes relative to the housekeeping gene Rps17. Data pooled from three experiments, Pou2f3−/− RQ normalized to average of WT mice in each experiment. Statistical significance determined by unpaired student’s T-test.
