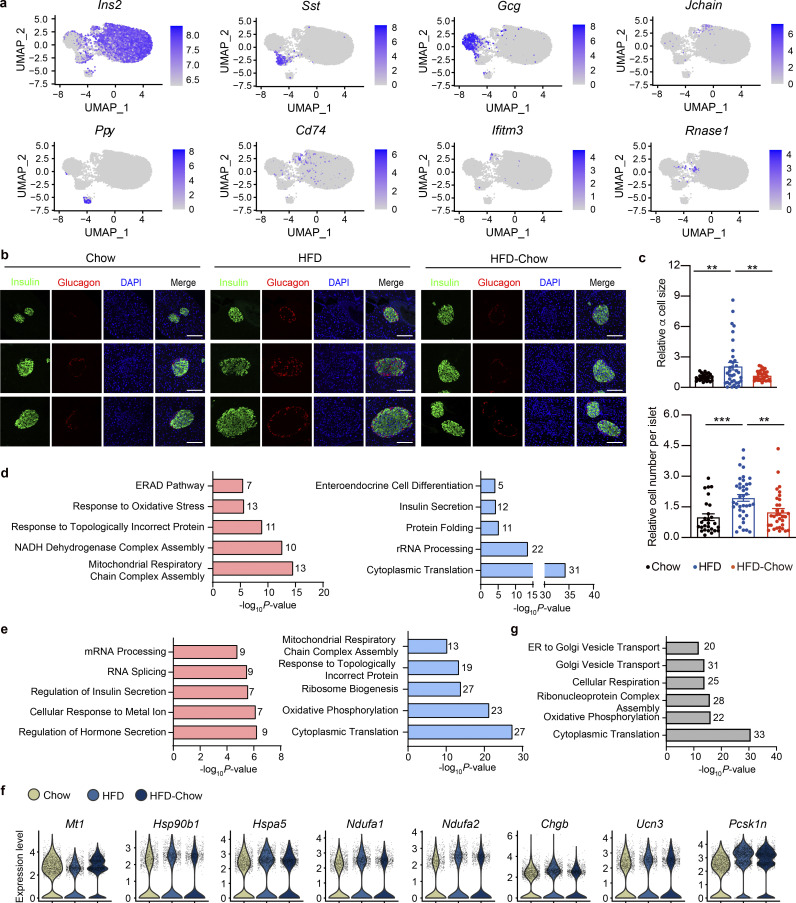
Figure S3.
Additional details of scRNA-Seq analysis. (a) scRNA-Seq and UMAP visualization of islet single cells from Chow, HFD, and HFD-Chow groups. Expression levels of indicated marker genes are shown. Scale bar indicates gene expression level. (b) Representative immunofluorescent images of insulin (marker for β cell) and glucagon (marker for α cell) staining in pancreas sections from indicated groups. Scale bar, 100 μm. (c) Relative α cell size and islet cell number quantified from immunofluorescent images and normalized to the average α cell size, and the islet cell number, respectively, in the Chow group. Data represent mean ± SEM (n = 4–5 mice per group, dots represent islets); **, P < 0.01; ***, P < 0.001; one-way ANOVA. (d) GO biological process analysis of upregulated (left) and downregulated (right) genes in β cell clusters from HFD group compared to Chow group. (e) GO biological process analysis of upregulated (left) and downregulated (right) genes in β cell clusters from HFD-Chow group compared to HFD group. (f) Violin plot of selected gene expression, expression level in each cell was marked in gray dot. (g) GO biological process analysis of genes unaltered by dietary intervention in β cell clusters (P < 0.05 AND NOT Reversible Genes). In d, e, and g, most significant and nonredundant biological processes with respective gene numbers and P values are shown. Significantly changed genes were identified by P < 0.05 with Wilcoxon Rank Sum test. Data in b and c are representative of two independent experiments.