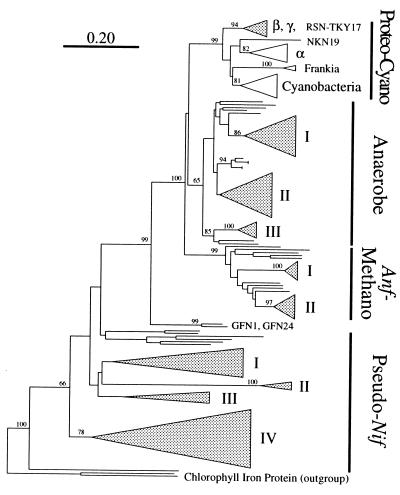
FIG. 2.
A large phylogenetic tree showing the relative positions of the major nifH groups and the major clusters of nifH genes isolated from the microbial communities of termite guts. The tree was constructed by the neighbor-joining method, and bootstrap values above 50 from 100 resamplings are shown for each node. Chlorophyll iron proteins were used as outgroups. The scale bar denotes 0.20 substitutions per site. Shaded wedges indicate the clusters consisting of sequences derived from termites. The depths and widths of the wedges reflect the branching lengths and the numbers of clones within the clusters, respectively. The proteo-cyano group includes conventional nifH sequences from proteobacterial clades (α, β, and γ), Frankia spp., and cyanobacteria. The anf-methano group represents anfH genes of molybdenum- and vanadium-independent nitrogenases and functional molybdenum-dependent nitrogenase genes from methanogenic archaea. The anaerobe group includes sequences from (low G+C gram-positive) clostridia, sulfate reducers (δ-proteobacteria), and M. barkeri (methanogenic Archaea domain). The pseudo nif group includes sequences from divergent genes of methanogens that might not encode active nitrogenases. The roman numbers indicate the clusters consisting of the sequences from termites in each group (Fig. 3 to 5 and see text).