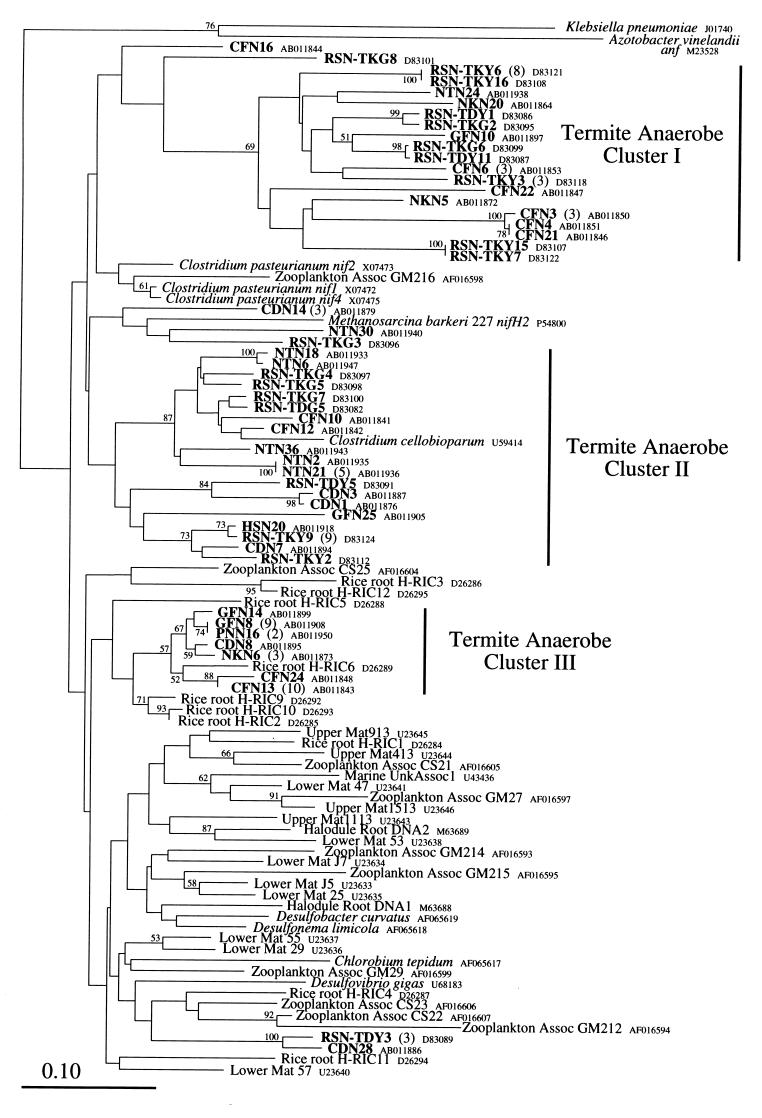
FIG. 3.
Phylogenetic relationships of nifH sequences within the anaerobe group. The tree was constructed by the neighbor-joining method based on 90-amino-acid alignment positions corresponding to positions 45 to 129 in the K. pneumoniae nifH protein. Bootstrap values above 50 from 100 resamplings are shown for each node. The sequences of K. pneumoniae and Azotobacter vinelandii anfH were used as outgroups. The scale bar shows 0.10 substitutions per site. Three clusters (I, II, and III) consisting of sequences from termites are indicated. Numbers in parentheses denote numbers of clones having identical amino acid sequences from a single termite species (clones with unique sequences are not shown).