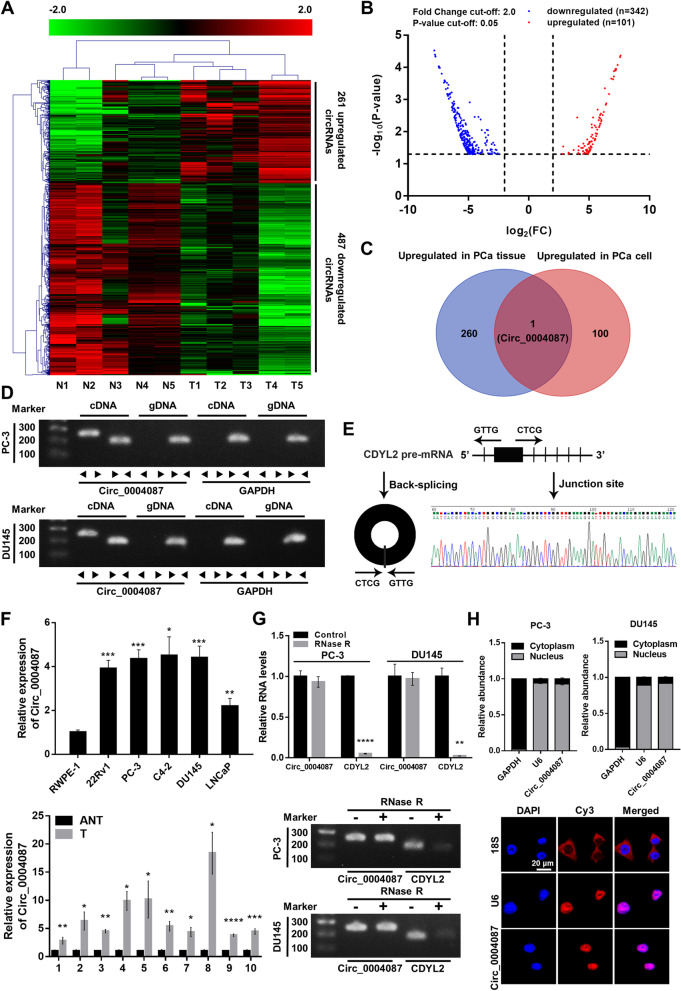
Fig. 1.
Circ_0004087 is significantly upregulated in PCa. (A) The heatmap of circRNA expression pattern in PCa tissue generated from microarray profile. (B) The differentially expressed circRNAs in PCa cell line identified by RNA-seq data. |log2(FC)|≥ 2, P < 0.05. (C) The venn diagram showing the intersection of the microarray profile and the RNA-seq data. (D) The existence of circ_0004087 in cDNA or genomic DNA (gDNA) of PCa cell lines determined using divergent and convergent primers. GAPDH was used as a negative control. (E) Schematic illustration showing the genomic location and the junction site of circ_0004087. (F) (Top) Relative levels of circ_0004087 in human prostatic epithelial cell line (RWPE-1) and PCa cell lines (22Rv1, PC-3, C4-2, DU145, and LNCaP). (Bottom) Relative levels of circ_0004087 in 10 pairs of PCa and adjacent normal tissues. ANT, Adjacent normal tissues; T, Tumor tissues. (G) The expression of circ_0004087 and its linear counterpart CDYL2 after RNase R treatment in PC-3 and DU145 cells. (H) RNA nucleus/cytoplasm separation assay (top) and RNA-FISH assay (bottom) revealing the distribution of circ_0004087 in PCa cell. 18S and GAPDH were applied as positive controls in cytoplasm, U6 was applied as a positive control in nucleus. Circ_0004087, 18S, and U6 probes were labeled with Cy3, nuclei were stained with DAPI. 1600 × . Statistical analysis is shown on the bar graphs. Data are presented as the mean ± SD of three independent experiments. * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001