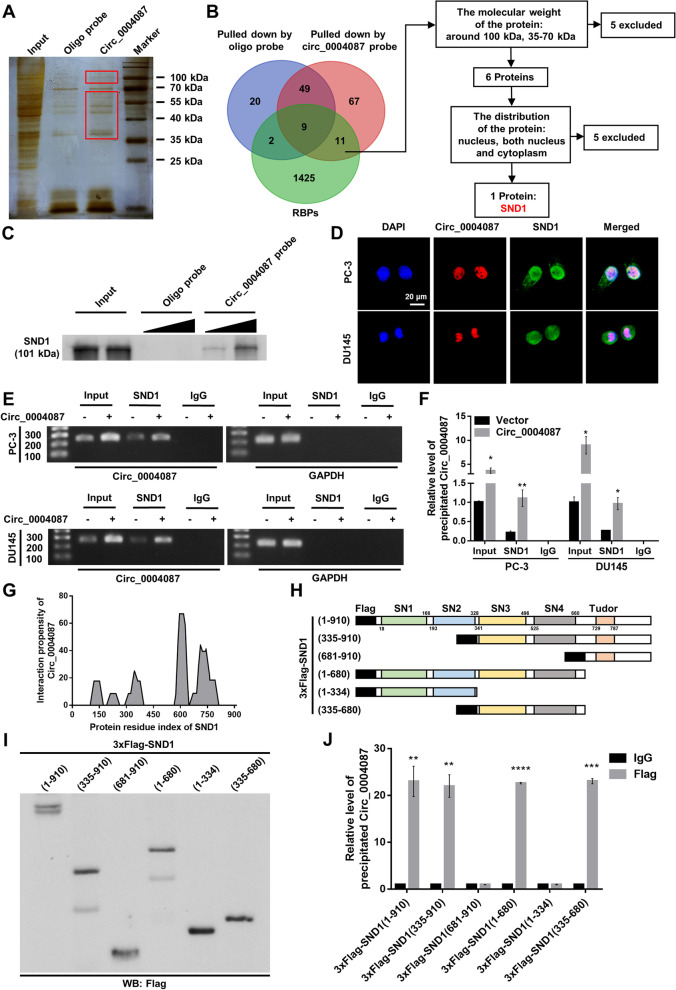
Fig. 3.
Circ_0004087 interacts with SND1 in PCa cells. (A) Silver staining showing proteins that interact with circ_0004087 in PCa cells. Red boxes show the location of differential bands. (B) The flow chart for screening out the protein partner of circ_0004087. (C) Western blot verifying the protein pulled down by biotin-labeled circ_0004087 probe. (D) RNA-FISH and immunofluorescence staining assays exhibiting the subcellular localization of circ_0004087 (red) and SND1 (green) in PCa cell lines. Nuclei were stained with DAPI 1600 × . (E) The result of RIP assay with anti-SND1 antibody showed by agarose gel electrophoresis diagram. IgG was used as a negative control. (F) Quantification of the result of RIP assay with anti-SND1 antibody. IgG was used as a negative control. (G) The binding propensity between circ_0004087 and SND1 protein predicted by the catRAPID algorithm. (H) Structure diagrams showing full-length and truncated Flag-tagged recombinant SND1 proteins. (I) Western blot validating the full-length and truncated Flag-tagged recombinant SND1 proteins. (J) RIP assay with anti-Flag antibody revealing the binding domains of SND1 to circ_0004087. IgG was used as a negative control. Statistical analysis is shown on the bar graphs. Data are presented as the mean ± SD of three independent experiments. * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001