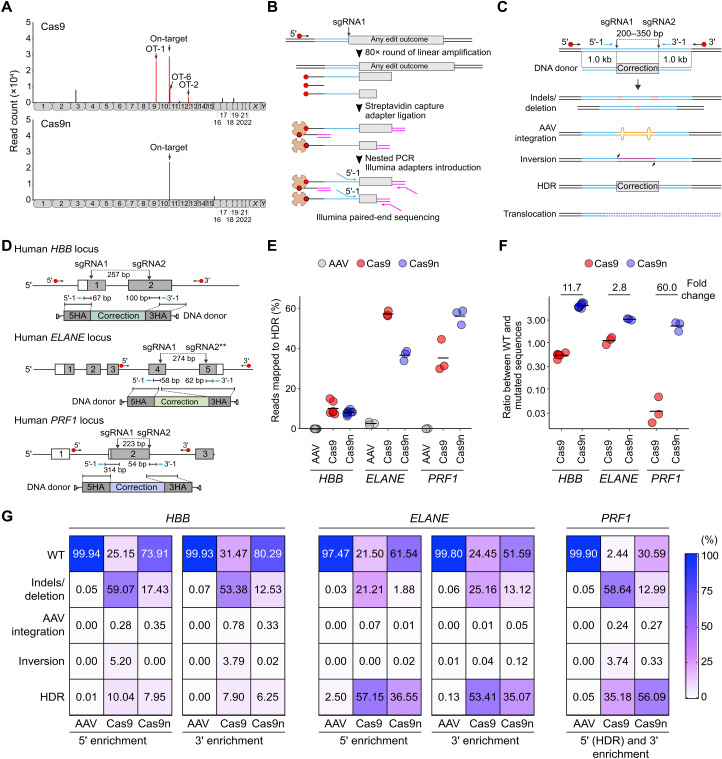
Fig. 5. Analysis of AAV integrations and complex genetic alterations by AAV-seq and LAM-HTGTS.
(A) AAV integration sites identified and mapped on human chromosomes of HBB-targeted HSPCs treated with sgRNAs/Cas9 (Cas9) or sgRNAs/Cas9n (Cas9n) RNPs and AAV donor vectors. The top hits of AAV integrations are indicated in red, including the on-target site and three high-risk off-target sites (OT-1, OT-2, and OT-6), as identified by GUIDE-seq. Random AAV integrations are shown in black. (B) Scheme of LAM-HTGTS. Red circles depict biotin, and orange counterparts depict streptavidin. (C) Scheme of the spacer-nick gene repair approach (top) and all the possible genetic outcomes including small indels/and deletion, AAV integration, inversion, HDR events, and translocations. LAM-PCR was performed using external biotinylated primers annealing outside of the 5′ or 3′ HAs and nested primers nearby the first cleavage site (5′-1 or 3′-1). (D) Schematic representation of the targeted loci analyzed by LAM-HTGTS. Two asterisks indicate that this sgRNA was not combined with Cas9. (E) HDR efficiencies detected by LAM-HTGTS at the targeted loci. (F) Ratio of WT:mutated sequences in the HSPCs treated as indicated. Data are shown as means ± SD from at least three independent experiments. (G) Heatmaps of the frequencies of all outcomes depicted for both 5′ and 3′ enrichments at the targeted HBB and ELANE loci or combined enrichment (5′ and 3′) at the targeted PRF1.