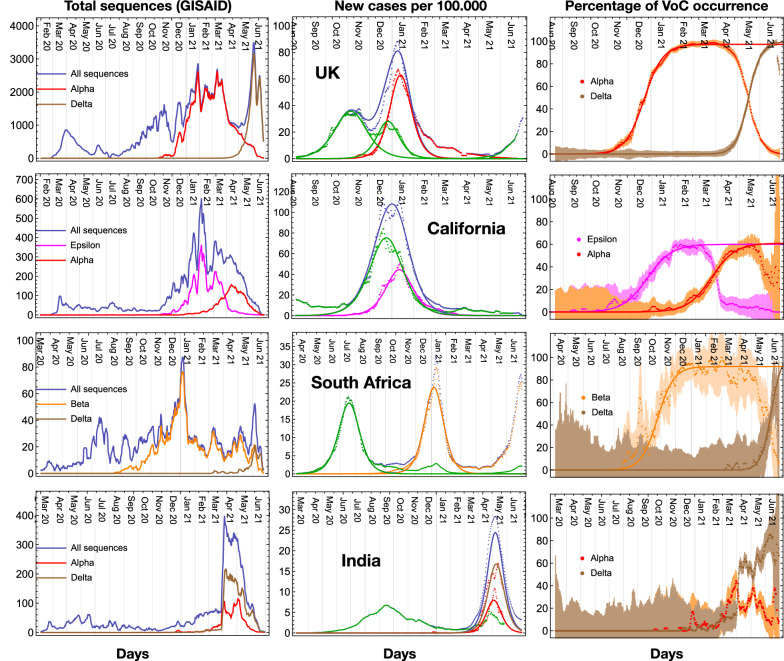
Figure 6.
MeRG model for epidemiological data of variants. Results of the MeRG fitting of the number of infected associated to each relevant variant. Each row corresponds to a geographical region. In the left column we show the total number of sequencing available on GISAID (in colour the ones associated to the relevant VoC or VoI); the middle column shows the number of new daily infected (per 100.000 inhabitants); the right column shows the percentage of each VoC or VoI in the sequencing data. All plots show daily rates, with data smoothened over a period of 7 days. In the middle plots, the data are shown by dots, where blue corresponds to the total and the colours show the number of infected associated to each variant. The solid lines show the result of the fits to the MeRG model (note that only for the UK we fit the “standard variant”—in green-with two logistic functions). In the left plots, the error derives from the expected statistical variation on the number of daily sequences (after smoothening). For all the plots, the classification in variants derived from the GISAID data.