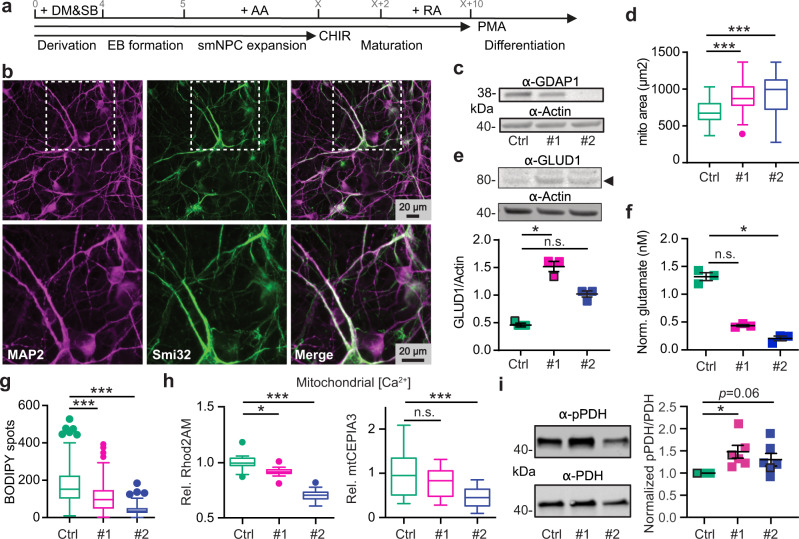
Fig. 4. Patient-derived cells are similarly characterized by an anaplerotic state and reduced mitochondrial Ca2+ levels.
a Differentiation protocol to obtain motoneurons (MN) from induced-pluripotent stem cells (iPSCs) and (b) confirmatory immunocytochemistry by staining against dendrite marker MAP2 and motoneuronal neurofilament Smi32. c Immunoblot of total MN cell lysates for validation of GDAP1 expression, actin served as loading control, size is indicated. d Automated high-content microscopy analysis of MitoTracker-stained mitochondria revealed an increased total mitochondrial mass per well. e Immunoblot of total MN cell lysates demonstrated increased glutamate dehydrogenase (GLUD1, arrowhead) in patient-derived MNs, actin served as loading control. f Reduced glutamate levels in CMT4A patient-derived MN determined by a luminescence-based assay indicating increased glutamine consumption compared to Ctrl. g Automated high-content microscopy analysis of BODIPY-positive spots normalized to the nuclei area in MN revealed a reduced number of lipid droplets in CMT4A patient-derived cells. h Reduced relative mt[Ca2+] levels of NPCs determined by confocal microscopy with Rhod2-AM and mito-CEPIA3. i Immunoblots and quantification demonstrating increased phosphorylation of PDH E1 serine 293, normalized to total PDH E1 levels and control cells. Data in (d–g) were from 3 independent differentiations and data in (d, g) were obtained from n > 40,000 cells in quadruplicate, experiments in (f) were performed in duplicate. Data in (h) were from 20–22 Rhod2-AM-stained cells and 22–33 cells mitoCEPIA3 transfected from three independent experiments. Data in (i) were from 6 different passages. Data points corresponding to the example blots are highlighted. Statistical variation is shown as Tukey boxplots in (d, e) or scatter plots in (i) with mean ± SEM indicated. Significance was calculated using the non-parametric Kruskal–Wallis test, *p < 0.05, **p < 0.001, ***p < 0.0001.