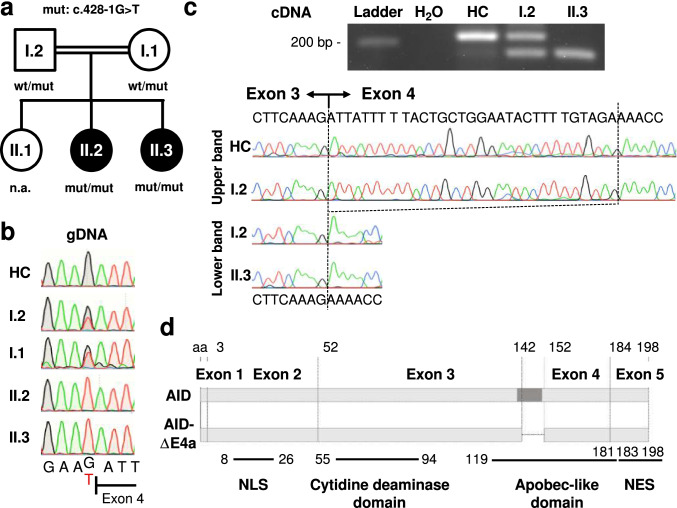
Fig. 1.
Identification of a homozygous AID-ΔE4a splice site variant in two patients with Hyper-IgM syndrome. a Pedigree of the affected sisters with clinical phenotype of hyper-IgM syndrome (II.2 and II.3). Allelic distribution of AICDA wild-type (wt) and the acceptor splice site variant (mut, c.428-1G > T) is indicated (n.a., not assessed). b Sequence chromatograms (gDNA) of the acceptor splice site before AICDA exon 4 in a healthy control (HC), the parents and both patients. Resulting nucleotide sequences are depicted below. c Gel electrophoresis of AICDA exon3/4 boundary RT-PCR products derived from CD40L/IL-21 stimulated B cells of a healthy control (HC), the affected patient (II.3) and the father (I.2). The chromatograms and resulting nucleotide sequences of the corresponding bands are depicted below. d Schematic representation of the AICDA exons and the 10 amino acid (aa) deletion in AID-ΔE4a. A DNA segment encoding a highly conserved alpha helix is indicated in darker grey [22]. Different functional AID domains are shown in the lower part [9] (NLS, nuclear localization signal; NES, nuclear export signal)