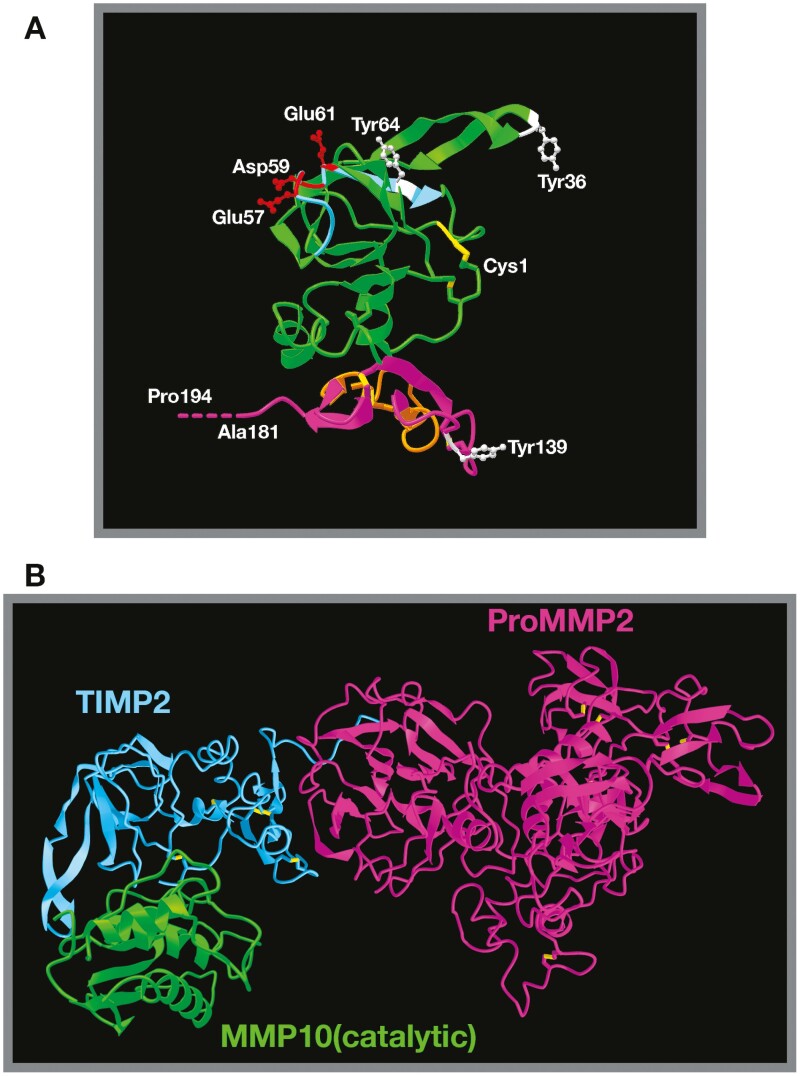
Figure 2.
Structure of TIMP2 alone and in complex with pro-MMP2/active MMPs.
(A) TIMP2 3D structure based on X-ray diffraction studies (Protein Data Bank structure 1BR9) (68) demonstrating the classic OB-fold structure. The N-terminal domain is highlighted in green, the C-terminal domain in purple. The six disulfide bonds critical for correct secondary structure are highlighted in yellow and the position of the three phosphorylated tyrosine residues (Tyr36, Tyr64 and Tyr139 of the mature secreted protein) are shown in white, with Tyr64 being critical for regulation of MMP2 and HSP90 interactions. The BC loop (light blue) containing three acidic amino acid residues (Blu57, Asp59 and Glu61, shown in red) is the proposed integrin binding domain. Also shown are the N-terminal cysteine (Cys1) and C-terminal alanine residues (truncated in the analysis). The true C-terminal residue is a proline residue (Pro194), which has been artificially extrapolated and labeled. The orange domain within the C-terminal domain highlights loop 6, which is responsible for direct antagonism at the IGF1-receptor. (B) Structure of TIMP2 in complex with pro-MMP2 (68) superimposed with TIMP2 complexed with the catalytic domain of MMP10 (12) (TIMP2 from this image is hidden) emphasizes the fact that TIMP2 proteinase inhibitory activity is mostly unhindered when in complex with pro-MMP2. The fate of this theoretical heterotrimer is uncertain.