Abstract
Purpose
Due to growing concerns about the obesity pandemic as a worldwide phenomenon, a global effort has been made for managing it and associated disorders. Accordingly, metabolomics as a promising field of “OMICS” is presented for investigating different molecular pathways in obesity and related disorders through the evaluation of specific metabolites in both animal and human subjects. Herein, the aim of the present study as the first systematic review is to evaluate all available studies about different mechanisms and their biomarkers discovery using metabolomics approaches.
Method
The study was designed according to Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines. Using a comprehensive search strategy we searched in databases including; Web of Science, PubMed, and Scopus using specific keywords. Based on predefined inclusion/exclusion criteria study selection has been conducted considering the type of studies, participant, and outcome measures. Quality assessment was done using CASP (Critical Appraisal Skills Programme) checklist followed by data extraction according to a predefined data extraction sheet.
Results
Among the articles that resulted from electronic search, a total of 74 articles met our inclusion criteria. The most prevalent studied metabolites were amino acids and lipid derivatives and both targeted and non-targeted approaches were applied for metabolomics studies.
Conclusion
This systematic review summarized a wide range of studies regardless of the age, history, language, and type of the study. Further studies are needed to compare the application of emerging methods in the treatment of obesity and related disorders.
Supplementary Information
The online version contains supplementary material available at 10.1007/s40200-021-00917-w.
Keywords: Metabolomics, Obesity, Metabolic syndrome, Metabolite
Introduction
Obesity as a widespread problem with a simultaneous increase in all around the world has great implication in public health. There is a growing concern about the obesity-associated disorders and related risks. Obesity is a pandemic disease of the present century by the World Health Organization (WHO) and other international organizations [1, 66]. Worldwide, obesity prevalence has been estimated by 1.5 billion people. According to the WHO, in 2016, over 1.9 billion (39%) individuals aged 18 years and older were overweight, among which more than 650 million (13%) were considered obese [20, 97]. Obesity is associated with the risk of several disorders especially chronic diseases such as; diabetes, cancer, metabolic syndrome, liver disease, cardiovascular disease [71, 75]. According to the WHO report, the obesity annual incidence is approximately 0.8–0.9% [53]. Globally, obesity is the cause of 5% of mortality and morbidity and its economic burden has been estimated $2 trillion [2]. The high prevalence can partially be attributed to the increasing consumption of hypercaloric, junk foods, and sedentary lifestyles [72, 73]. It is necessary to know the molecular pathogenesis of weight change for developing successful strategies for losing weight. Changes in the metabolomics profiles and described models can be used as an accurate predictor for obesity and obesity-related disorders [94]. Recently, a number of metabolites and biomarkers have been identified in different animal models of obesity and human subjects using metabolomics methods and metabolomic profile evaluation [13]. Metabolomics, a promising field of “OMICS”, is considered the best tool for metabolite and phenotype identification [86]. Metabolomics is a technological mechanism that can identify and measure variations in the profiles and levels of low molecular weight metabolites (< 1500 Da) in cells, tissues, organs, systems, or whole organisms in reply to a genetic variation, pathological or physiological state [31]. Therefore, metabolomics evaluates changes in metabolites due to obesity at the cellular level, i.e., visceral and omental white adipose tissues (AT), brown AT, skeletal muscle, and liver. Also, it can ascertain the metabolic fingerprinting (a determined chemical pattern special to an individual sample) relevant to metabolically unhealthy obese individuals compared to metabolically healthy individuals [8]. Metabolomics involves qualitative and quantitative analyses of intracellular and intercellular metabolites, usually using two main distinct analytical approaches including; a) nontargeted metabolite profiling (comprehensive analysis without further knowledge of the features which might result in the identification of a large variety of metabolites that can cluster into recognizable patterns). b) targeted metabolite profiling (focused on reliable quantitative measurement of the variations in metabolites involved in several metabolic pathways (e.g., amino acids (AA) and their derivatives) based on their biological roles in those pathways) [70]. These methods differ in various aspects, such as the complexity of sample preparation procedures, experimental precision, range of features (metabolites) identified, and the quantification level (relative versus absolute) [78]. Those features assist researchers to establish particular objectives for each approach, such as creating a hypothesis or testing an earlier developed hypothesis [77]. Metabolites are important molecular biomarkers for diagnosis and prognosis of different disorders. In other words, the role of these small molecules in biological systems is considerable and they are a suitable choice for the perception of obesity phenotypes. In recent decades, prevalence of obesity has a warning progressive rise rate in children, adolescents, and adults. Accordingly, understanding obesity mechanisms has great importance which leads to reduce burdens imposed by and improve patient health status and life quality [107]. Nevertheless, there are still a few studies that systematically review obesity and related biomarkers. In this respect, the aim of present study as the first systematic review of the relationship between obesity and metabolites is to evaluate all available studies about different mechanisms underlying obesity and its biomarkers discovery using metabolomics approaches. Specifically, this systematic review will be covering all relevant literature regardless of age, history, and language. Generally, results of this study, based on databases in this area, can be beneficial as valuable sources for future studies.
Materials and methods
Study design
In this article, the relationship between obesity and metabolites have been systematically reviewed. This systematic review protocol was registered in the International Prospective Register of Systematic Reviews (Registration number: CRD42018104857).
Search strategy and data collection
All studies about the association between metabolites and the profile of metabolite with obesity searched and reviewed. For this purpose, the databases, including Web of Science, PubMed, and Scopus were searched. The search algorithm was included all possible combinations of keywords from the following: “Metabolomics”, “Metabolome”, “ metabotropic quisqualate receptor “,” Metabolite Profiles”, ““MSAG protein” “obesity”, “weight”, “obese”, “body mass index”, and “ metabolic syndrome “ (Table 1). In addition to electronic resources, the national, regional, and international congresses were searched. Also, references of related review and systematic review articles were reviewed to increase coverage of included articles and ensure literature saturation. At least three emails with logical intervals (about 2 weeks) were sent to the corresponding author of the article in order to eliminate the limitations of no access to full text.
Table 1.
Search strategy
| PubMed | |
| (((((((“Metabolome”[Mesh]) OR “Metabolomics”[Mesh]) OR “metabotropic quisqualate receptor” [Supplementary Concept]) OR “Metabolite Profiles”) OR “MSAG protein, human” [Supplementary Concept]) AND Humans [Mesh])) AND ((((((“Obesity, Abdominal”[Mesh]) OR “Abdominal obesity metabolic syndrome” [Supplementary Concept])) OR obesity)) OR “Body Mass Index”[Mesh]) | |
| Scopus | |
| (TITLE-ABS-KEY (metabolom*) OR TITLE-ABS-KEY (metabotropic) OR TITLE-ABS-KEY (“Metabolite Profiles”) OR TITLE-ABS-KEY (“MSAG protein”) OR TITLE-ABS-KEY (“Metabolomics”))) AND ((TITLE-ABS-KEY (obesity) OR TITLE-ABS-KEY (“BMI”) OR TITLE-ABS-KEY (“Body Mass Index”) OR TITLE-ABS-KEY (“Body Weight”))) AND (LIMIT-TO (DOCTYPE, “ar”) OR LIMIT-TO (DOCTYPE, “re”)) AND (LIMIT-TO (SUBJAREA, “MEDI”) OR LIMIT-TO (SUBJAREA, “BIOC”)) AND (LIMIT-TO (SRCTYPE, “j”)) | |
| ISI/WOS | |
|
TOPIC: (metabolom*) OR TOPIC: (“Metabolite Profiles”) OR TOPIC: (“MSAG protein”) OR TOPIC: (metabotropic) Indexes = SCI-EXPANDED, SSCI, CPCI-S, CPCI-SSH, ESCI Timespan = All years TOPIC: (obes*) OR TOPIC: (“BMI”) OR TOPIC: (“Body Mass Index”) OR TOPIC: (“Body Weight”) Indexes = SCI-EXPANDED, SSCI, CPCI-S, CPCI-SSH, ESCI Timespan = All years #2 AND #1 Indexes = SCI-EXPANDED, SSCI, CPCI-S, CPCI-SSH, ESCI Timespan = All years |
Study selection criteria
Types of studies
The total of observational studies, including descriptive studies (cross-sectional, case control) and analytical studies (raw data of case studies and RCT and receiving placebo groups in these studies and cohort studies) that evaluate the association between metabolites and the profile of metabolite with obesity were recruited. If some disease or particular traits is influencing the dependent variable (obesity), these data not analyzed. All studies were independently screened by the review authors based on their titles and abstracts. The full text of potentially suitable articles was obtained to assess their relevancy based on the inclusion/exclusion criteria. Regardless of any language or date restriction, all related studies included. In this respect, the objective is access to studies that examine the relationship between metabolites and metabolite profiling with obesity.
Types of participants
Those studies evaluating the general adult human population (≥18 years) as well as child and adolescents participants (under 18 years of age) were included. They all have been conducted on overweight or obese individuals (body mass index [BMI] ≥ 25) or adults with metabolic syndrome (based on Adult Treatment Panel III and International Diabetes Federation criteria) while studies with populations restricted to specific conditions, diseases, or metabolic disorders were excluded.
Types of outcome measures
The outcomes are body weight, BMI, waist circumference, body fat, and metabolic syndrome.
Data extraction and quality assessment
The quality assessment of the included studies were assessed independently by two blind authors using Critical Appraisal Skills Programme (CASP) checklist.
Data were extracted independently from included studies by two authors according to a predefined data extraction sheet. Probable disagreements were resolved by discussion between the two authors, and consultation was made with a third author. Extracted data were including:
General information (author, publication year, type of study, study population, and location
Participants (sample size, sex, BMI and age)
Outcomes and main findings (reported outcomes: BMI, Body fat, waist circumference, and metabolic syndrome)
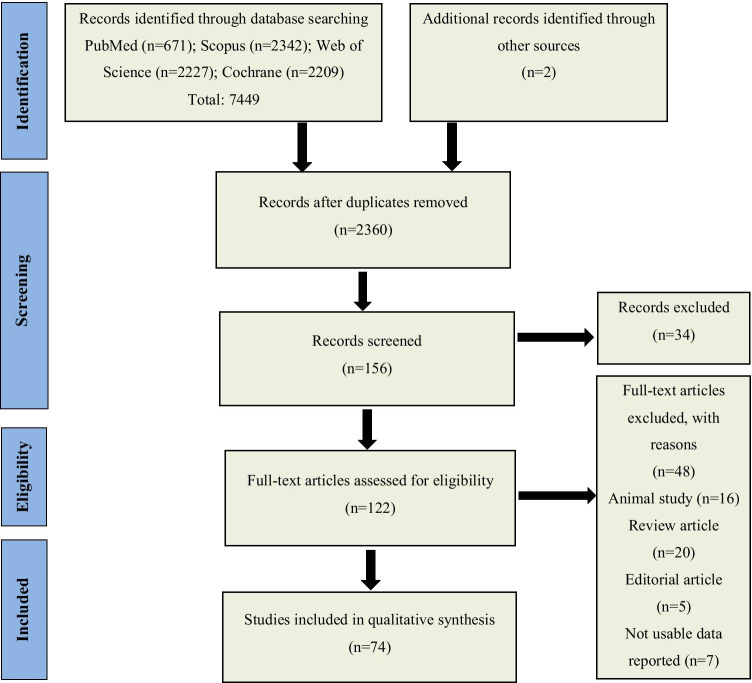
The whole process of study selection is summarized in the Preferred Reporting Items for.
Systematic Reviews and Meta-Analyses flow diagram PRISMA.
Ethical considerations
Proposals of the study were passed by the ethical committee of the EMRI (1396–03–111-2220). In this study, ethical approval is not essential because it is a secondary type of study and is not included, individuals. In fact, here results discussed through peer-reviewed publications (Fig. 1).
Fig. 1.
Flow chart for study identification and selection
Results
According to a comprehensive electronic search, 74 studies met our inclusion criteria. All articles extracted from mentioned databases were precisely evaluated based on the full text and reported supplementary data. A summary of the final extracted data from included articles is represented in Table 2.
Table 2.
Summary of the final extracted data
| ID | Author, Y | Country | Geographical Expansion national/subnational/international | Sample Size | Sex | Age | BMI | Sample | Method | Design | Score |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Lokhov, P.G, 2020 [58] | Russia | Local | 100 |
F & M |
Normal: 31.3 _ 5.5 Overweight: 32.9 _ 6.7 Stage 1 obesity: 29.7 _ 8.0 Stage 2 obesity: 32.8 _ 8.1 Stage 3 obesity: 34.5 _ 6.5 |
Normal: 22.1 ± 1.9 Overweight: 27.5 ± 1.3 Stage 1 obesity: 32.5 ± 11.7 Stage 2 obesity: 36.9 ± 1.3 Stage 3 obesity: 47.3 ± 6.1 |
Plasma | Mass Spectroscopy | Cohort study | 7 |
| 2 | Chashmniam, S, 2019 [17] | Iran | Local | 86 |
F & M |
Obes:23–35 | Obese: BMI ≥30 kg/m2 | Serum | H-NMR | Case-control study | 7 |
| Non-obese: 24–30 | Non-obese: BMI < 30 kg/m2 | ||||||||||
| 3 | Troisi, J, 2019 [91] | Italy | Local | 41 |
F & M |
7–15 years |
Obese: BMI > 95th percentile |
Saliva |
PLS-DA GC-MS |
Pilot-nested case-control study | 6 |
|
Normal Weight: (BMI) < 85th percentile | |||||||||||
| 4 | Shokry, E, 2019 [88] | Germany | Local | 325 |
F & newborn |
NW: 31.00 ± 6.00 | NW: 21.87 ± 2.66 |
Cord Plasma Cord Blood |
LC–MS/MS FIA–MS/MS |
PREOBE study, A prospective observational cohort study |
6 |
| Obese: 31.00 ± 4.75 | Obese: 28.83 ± 4.31 | ||||||||||
| 5 | Kim, M. J, 2019 [47] | Korea | Local | 77 | F | middle-aged |
low-BMI (n = 40, BMI <23 kg/m2), and high-BMI (n = 37, BMI >23 kg/m2) groups |
Plasma Proteins |
UPLC-Q-TOF -MS |
Genome-wide association study | 7 |
| 6 | Hsu, Y. H, 2019 [38] | United States of America | Local | 298 | F | 38.5 ± 12.1 | 28.3 ± 9.95 | Plasma |
LC-MS PAIRUP-MS |
Cohort study | 8 |
| 7 | Hellmuth, C, 2019 [35] | Germany | Local |
253 F |
F | 29 | 25.83 [8.37] [kg cm − 2] | Plasma | Whole-body dual X-ray absorptiometry (DXA) | Cohort study | 8 |
|
121 new born (M) |
New born | 3 | 12.88 (7.97) | ||||||||
| 8 | Feng, R, 2019 [27] | China | Local |
60 NW: 30 Obese: 30 |
M | 19 to 25 years of age |
Obese: (BMI) ≥ 28.0 kg/m2 |
Urine | UPLCQ-TOF MS | A cross-sectional study | 9 |
|
Normal weight: 18.5 kg/m2 < BMI < 24 kg/m2 | |||||||||||
| 9 | Bagheri, M, 2019 [12] | Iran | National |
300 NW: 100 Obese: 213 |
F & M |
Tween 18 and 50 years |
Obese: BMI ≥ 30 kg/m2 |
Plasma | LC-MS/MS | Case-control study | 8 |
|
NW: (18.5 ≤ BMI < 25 kg/m2 |
7 | ||||||||||
| 10 | Yu, H. T, 2018 [106] | China | Local | Obese: 36 | M |
Obese: 22.7 ± 2.25 |
Obese: (BMI) ≥25 kg/m2 |
Blood Urine |
UPLC-Q-TOF-MS | Case control study | 7 |
| NW: 35 | NW: 22.7 ± 2.50 |
NW: (18.5 kg/m2 ≤ BMI ≤ 22.9 kg/m2) |
|||||||||
| 11 | Xia, B, 2018 [103] | China | Local |
Obese: 69 NW: 80 |
F & M |
10–12: 29 (36.25) 13–15: 51 (63.75) |
Obese: 24.69 ± 2.94 |
Urine |
ESI-MS/MS HPLC GC-MS |
Case-control study | 8 |
| NW: 17.84 ± 2.25 | |||||||||||
| 12 | Wijayatunga, N. N, 2018 [101] | USA | Local | 20 |
F & M |
Pre surgery: 37.25 (11.68) 2 weeks: 37.60 (11.07) 6 months: 37.62(12.92) |
Pre surgery: 46.83 (6.21) 2 weeks: 43.65 (6.42) 6 months: 4.34 (6.44) |
Serum | NMR | Pilot study | 8 |
| 13 | Wang, S. M, 2018 [100] | China | Local |
600: 328 men and 272 women Obese: 302 NW: 298 |
F & M |
Obese: 66 ± 11 |
Obese: BMI ≥ 24.0 kg m−2 | Serum |
LC-MS/MS HPLC |
Cross-sectional study | 8 |
| NW: 62 ± 17 | NW: 18.5 < BMI < 24.0 kg m−2 | ||||||||||
| 14 | Seridi, L, 2018 [87] | USA | Local |
27 Obese:18 |
F | Obese:; BMI > 35 kg/m2 | Plasma | PLS-DA |
Cohort Study |
7 | |
| NW: 9 |
NW: (BMI) < 25 kg/m2 |
||||||||||
| 15 | Romo-Hualde, A, 2018 [81] | Spain | Local | 70 | F | Obese: 37.3 ± 7.6 years old | Obese: 31.6 ± 3.1 BMI | Urine |
HPLC-TOF-MS LC-MS |
A double blind randomized placebo-controlled intervention study | 7 |
| NW: 39.0 ± 8.0 | |||||||||||
| 16 | Palmnas, M. S. A, 2018 [68] | Canada | Local |
N = 82 Men:35 Women:47 |
F & M |
aged 30–60 years |
Obese women: 24.6 (2.8) Obese men: 27.4 (2.5) |
Serum | STAR-Q | Systematic study | 6 |
|
NW women: 21.6 (2.0) Men: 23.4 (1.9) |
|||||||||||
| 17 | Palau-Rodriguez, M, 2018 [67] | Spain | Local | 39 |
F & M |
Obesity:MH:39.29 ± 8.87 MU:42.56 ± 10.94 |
Obese: MH: 48.81 ± 9.12 MU: 52.51 ± 7.14 kg/m2 |
QC1 (Milli-Q Water Samples), QC2 (Aqueous Solution Of A Standard) |
sPLS-DA PCA ESI |
Systematic study | 9 |
| 18 | Leal-Witt, M. J, 2018 [54] | Spain | Local | 35 |
Children F & M |
7–10 years 8.9 (8.6–9.3) |
3.56 (3.29–3.84) | Plasma |
LC-MS PCA |
Observational longitudinal study | 9 |
| 3.11 (2.88–3.34) | |||||||||||
| 19 | Bagheri, M, 2018 [10] | Iran | National | NWMH (n = 78) |
F & M |
NWMH: (Male): 33.5 (30–39.75) (Female): 36 (30.5–41.25) |
NWMH: (Male): 23.49 (22.19–24.72) (Female): 22.93 (21.52–24.09) |
Plasma |
LC − MS/MS Kruskal-Wallis test Wilcoxon’s Signed Rank test |
Case-control study | 8 |
|
Obese: MHO (n = 107) MUHO (n = 100) |
Obese: MHO: (male): 33 (30.5–39) (Female): 35 (30.75–42) MUHO: (male): 35 (29–39) (female): 37 (34–43) |
Obese: MHO: (male): 33.72 (31.92–36.54) (Female 34.32 (31.74–36.2) MUHO: (male): 34.78 (32.89–38.14) (female): 35.19 (32.17–39.12) |
|||||||||
| 20 | Bagheri, M, 2018 [10] | Iran | National |
MHO: 82 MUHO: 78 |
F & M |
Obese: MHO: (Placebo): 37.17 ± 7.11 (vitamin D): 37.077 ± 7.50 MUHO: (Placebo): 35.70 ± 7.99 (vitamin D): 35.08 ± 7.55 |
Obese: MHO: (Placebo): 33.94 (32.03–35.81) (vitamin D): 34.52 (31.84–36.89) MUHO: (Placebo): 0.405 33.6 (32.14–38.52) (vitamin D): 35.18 (33.18–38.08) (kg/m2) |
Plasma |
LC-MS/MS HPLC |
Two randomized clinical trials | 7 |
| 21 | Almanza-Aguilera, E, 2018 [5] | Spain | Local | 115 | F | Control: 44.4 ± 3.31 |
Baseline(Control): 36.3 ± 5.74 3 months: (control) 88.3 ± 13.8 12 months: (control) 86.7 ± 13.5 |
Plasma | H-NMR | Lifestyle weight loss (LWL) intervention study | 7 |
| Treatment: 45.7 ± 3.51 |
Baseline(treatment): 35.4 ± 4.12 3 months: (treatment): 31.7 ± 3.67 12 months: (treatment): 31.3 ± 4.19 |
||||||||||
| 22 | Sun, L, 2017 [90] | China | Local | 611 |
F & M |
Adults: age < 75 years 1.58 (1.21,2.05) Oldest-old: age > 85 years 1.25 (0.80, 1.94) |
BMI < 25 kg/m2: 1.02 (0.66, 1.58) BMI > =25 kg/m2: 1.76 (1.18, 2.63) |
Serum | LC/MS/MS | A long-term randomized study | 6 |
|
Tertile of serum BCAA: Low: 204 Middle:203 High:204 |
Low: 75.3 ± 23.1 Middle: 64.5 ± 20.2 High: 62.6 ± 20.6 |
Low: 20.6 ± 3.5 Middle: 22.8 ± 3.7 High: 23.8 ± 3.6 |
|||||||||
| 23 | Sallese, A, 2017 [83] | USA | Local |
Obese non-mets (n = 43) Obese mets (n = 26) |
F & M |
Age 65 years |
Obese non mets and obese mets groups BMI (35.2 ± 6.8 vs 35.6 ± 4.5) |
Serum | MS-based metabolomics | Pilot study | 7 |
| 24 | Fattuoni C, 2017 [26] | Italy | local (Milan) | 56 | F | 33.9 ± 5.2 | Obese: 36.4 ± 4.8 | Placenta Tissue | GC-MS | Case/control | 6 |
|
NW: 33.7 ± 5.7 |
Normal weight: 21.5 ± 1.6 | ||||||||||
| 25 | Zhong F, 2017 [109] | USA | local | 69 |
F & M |
29.3 ± 10.3 | Obese (BMI) 30 kg/m2 | Plasma | Targeted HPLC-MS/MS | Case/control | 7 |
| 27.4 ± 9.8 | |||||||||||
| 26 | Sandler V, 2017 [84] | USA | International European-ancestry | 400 mother–offspring dyads | F | ND | ND | Plasma | Targeted MS-based & non-targeted GC/MS | Case/control | 7 |
| 27 | Isherwood CM, 2017 [42] | Surrey | local | 23 | M | Lean = 53.6 ± 6.0 | ND | Serum | UPLC–triple quadrupole mass spectrometry, UPLC quadrupole time-of-flight mass spectrometry | Case/control | 7 |
| OW/OB = 51.0 ± 7.7 | |||||||||||
| T2DM = 57.3 ± 4.8 | |||||||||||
| 28 | Schlecht I, 2017 [85] | Germany | local | 228 |
F & M |
Total: 51.96 (12.55) | Total:26.61 (4.66) | Urine And Serum | NMR | Case/control | 8 |
| F:52.80 (12.00) | F:25.97 (4.99) | ||||||||||
| M:50.97 (13.15) | M:27.36 (4.13) | ||||||||||
| 29 | [56] [56] | China | National | 343 |
F & M |
N: 37.74 ± 0.84 | Healthy: 21.11 ± 0.13 | Serum | UPLC–triple quadrupole mass spectrometry, UPLC quadrupole time-of-flight mass spectrometry | Case/control | 8 |
| OW/OB39.09 ± 1.32 | OW/OB:26.72 ± 0.25 | ||||||||||
| OW/OB DM:57.41 ± 0.85 | OW/OB DM:27.82 ± 0.33 | ||||||||||
| 30 | Okekunle AP, 2017 [65] | China | Local | 200 |
F & M |
Serum | UPLC-TQ-MS | Cross sectional | 8 | ||
| Healthy controls:46.24 ± 8.48 | Healthy controls: BMI = 18–24 | ||||||||||
| Obese controls: 42.92 ± 12.37 | Obese controls: BMI ≥ 28 | ||||||||||
| MetS: 45.30 ± 11.43 | MetS: BMI ≥ 28 | ||||||||||
| 31 | Baek S H, 2017 [9] | Korea | local | LFO group (n 5 34) |
F & M |
30 to 65 | HFO: 25 ≤ BMI < 30 HFO & (VFA) at L4 ≥ 100 cm2] | Plasma | UPLC-LTQ-Orbitrap XL MS | Case/control | 8 |
| HFO group (n 5 34) | (LFO):controls 25 ≤ BMI < 30 & VFA at L4 < 100 cm2 | ||||||||||
| 32 | Hellmuth C, 2017 [34] | Germany | local: Bad Honnef & Munich | 753 children |
F & M |
ND | ND | Serum Of Venous Cord Blood | liquid chromatography-tandem mass spectrometry | Cohort | 9 |
| 33 | Murphy RA, 2016 [63] | British Columbia | International | 319 | black men | 72 (2.4) | 26.8 (23.8–30.0) | Plasma | LC-MS, Nexera X2 U-HPLC, Exactive Plus orbitrap mass spectrometer | Case/control | 7 |
| 34 | Ahmad MS, 2016 [3] | Saudi Arabia | Local | 98 |
F & M |
18 to 39 | Normal (18.50–24.99 kg/m2) | Urine, Serum | NMR | Case/control | 7 |
| Obese class I (30.00–34.99 kg/m2) | |||||||||||
| Obese class Il, (35.00–39.99 kg/m2) | |||||||||||
| Obese class lll, (≥40.00 kg/m2) | |||||||||||
| 35 | [92] [92] | Spain | Local | 64 |
F & M |
Control: 47 ± 15 | Non-obese if: BMI = 18,5–26,9 kg/m2; | Serum | LC- and FIAESI-MS/MS | Case/control | 8 |
| Case: 43.67 ± 11.30 | Morbidly obese if: BMI N 40 kg/m2) | ||||||||||
| 36 | [79] [79] | Australia | Local | 1011 |
F & M |
20 | ND | Plasma | Flow-injection mass spectrometry | Case/control | 8 |
| 37 | Menni C, 2016 [60] | UK | Local | 2401 | F | 56.91 (11.57) | 26.30 (4.90) | Plasma | Ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry | Case/control | 9 |
| 38 | Kim Y J, 2016 [46] | Korea | Local | 2577 |
F & M |
Carrier type (TA/AA): 57.48 ± 9.11 | ND | Serum | Liquid chromatography and flow injection analysis mass spectrometry | Case/control | 8 |
| Wild type (TT): 56.97 ± 9.03 | |||||||||||
| 39 | Iida M, 2016 [41] | Japan | Local | Original study population (n = 594) | F | 35 to 74 | Average BMI 23 | Plasma | Capillary electrophoresis-mass spectrometry | Case/control | 7 |
| Replication population (n = 283) | |||||||||||
| 40 | Hellmuth C,2016 [33] | California, Irvine | Local | 167 non-diabetic women | F | 27.7 (5.4) | 25.9 (6.0) | Plasma | LC-MS and flow-injected mass spectrometry | Cohort | 9 |
| 41 | Yin X, 2016 [105] | USA | Local | 82 |
F & M |
53 ± 12 | ≥30 kg/m2 | Serum | GC-MS | Cross sectional | 8 |
| 42 | Yin X, 2016 (Replicate) [105] | USA | Local | 84 |
F & M |
67 ± 6 | ≥30 kg/m2 | Serum | GC-MS | Cross sectional | 8 |
| 43 | Zhao Q, 2016 [108] | USA | Local | 431 |
F & M |
Normal:28.58 ± 13.59 | Normal weight (BMI < 25 kg/m2), | Plasma | LC-MS | Case/control | 8 |
| Overweight: 36.99 ± 12.24 | Overweight (25 kg/m2 BMI < 30 kg/m2) | ||||||||||
| Obese: 34.53 ± 13.25 | Obesity (BMI 30 kg/m2) | ||||||||||
| 44 | Dugas, L, 2016 [23] | Africa | International | 2500 | F | 24–45 | > 30 kg/m2 | Plasma | GC-TOF/MS | Cohort | 9 |
| 45 | Allam-Ndoul, B, 2016 [4] | France | National | 664 |
F & M |
18 and 55 | 25 kg/m2 | Blood | GC MS | Cohort | 10 |
| 46 | Ho JE, 2016 [36] | United States | National | 1264 | F | 55 | 27.5 ± 4.9 | Plasma | Mass spectrometry | Cohort | 8 |
| 47 | Wang Y, 2015 [99] | China | Local | 60 | M | 20 to 55 | Obese hyperlipemia (n = 30) BMI ≥28.0 kg/m2 | Serum | UHPLC-Q-TOF MS/MS | Case/control | 8 |
| Normal-weight (n = 30, 18.5 kg/m2 < BMI < 24 kg/m2) | |||||||||||
| 48 | Paris, D, 2015 [69] | Italy | National | 60 |
F & M |
Lean: 37.5 | Lean: 20.6 kg/m2 | Ebc | NMR | Case/control | 6 |
| Obes: 39.2 | Obes: 45.3 kg/m2 | ||||||||||
| 49 | Desert, R, 2015 [22] | France | National | 65 | F | overweight: 30.77 | overweight: 25–30 | Urine And Cord-Blood | NMR | Cohort | 7 |
| obese: 29.92 | obese: > 30 | ||||||||||
| 50 | Chen, H, 2015 [13] | Taiwan | National | 68 |
F & M |
32–34 | >25 kg m − 2 | Plasma Samples | LC-MS and GC-MS | Case/control | 8 |
| 51 | Würtz P, 2014 [102] | Finland | International | 12,664 |
F & M |
NFBC86 = 16 | NFBC86 = 21.2(3.4) | Serum | NMR | Cohort | 9 |
| NFBC66 = 31 | NFBC66 = 24.6(4.0) | ||||||||||
| YFS = 24–39 | YFS = 25.0(4.4) | ||||||||||
| FINRISK = 24–39 | FINRISK = 24.7(4.0) | ||||||||||
| 52 | Lin Z, 2014 [57] | China | Local | 163 |
F & M |
25–70 | BMI ≥ 25.0 | Serum | GC/MS | Case/control | 9 |
| 53 | Badoud F, 2014 [7] | Canada | National | 30 |
F & M |
35 ≤ Age ≤ 70 | Lean (male): BMI ≤ 28 | Serum And Adipose Tissue | GC/MS | Cohort | 8 |
| Lean (female): BMI ≤ 24 | |||||||||||
| Obese (male): BMI ≥ 28 | |||||||||||
| Obes (female): BMI ≥ 24 | |||||||||||
| 54 | Valcárcel B, 2014 [93] | UK | International | 7255 |
F & M |
NFBC1966 = non-obese: 18.5 ≤ BMI ≤25 and obese: BMI ≥30 | Serum | NMR | Cohort | 9 | |
| NFBC1986 (male) = non-obese: 17.0 ≤ BMI ≤24.2 and obese: BMI ≥28.2 NFBC1986 (female) = non-obese: 17.4 ≤ BMI ≤24.05 and obese: BMI ≥27.5 | |||||||||||
| 55 | Xie G, 2014 [104] | China | International | 388 |
F & M |
Healthy obese1: 23–64.5 Healthy lean1: 20.2–63.9 |
Healthy obese1: 25.0–32.5 Healthy lean1: 19.1–22.2 |
Serum | UPLC-QTOFMS & GC-TOFMS | Cohort | 9 |
|
Healthy obese2: 18–64 Healthy lean2: 15–65 |
Healthy obese2: 24.4–31.6 Healthy lean2: 17.4–21.6 |
||||||||||
|
Healthy obese3: 44–83 Healthy lean3: 41–81 |
Healthy obese3: 27–52.7 Healthy lean3: 21–24.8 | ||||||||||
| 56 | Dunn WB, 2014 [24] | UK | National | 1200 |
F & M |
19–81 | 25.63 | Serum | GC-MS & UPLC-MS | Cohort | 9 |
| 57 | Newbern D, 2014 [64] | U.S | International | 82 |
F & M |
12 to 18 | ≥ 85th | Blood | Beckman-Coulter clinical analyzer | Cohort | 10 |
| 58 | Huang CF, 2013 [39] | Taiwan | National | 99 |
F & M |
69 ± 13 | Normal <24 Over weight ≥ 24 | Urine | LC-MS | Case/control | 5 |
| 59 | Jourdan C, 2012 [44] | Augsburg in Southern Germany | Local | 965 |
F & M |
Obese = 274 | Serum | ESI-(LC-) MS/MS | Cross sectional | 4 | |
| Nonobes = 691 | |||||||||||
| 60 | Wang C, 2011 [98] | China | Local | 103 | M | Obese: 20.8 ± 1.8 | Obese: 32.0 ± 3.8 | Urine | UPLC/Q-TOF MS | Case/control | 8 |
| Normal weight: 21.4 ± 2.0 | Normal weight: 20.6 ± 1.5 | ||||||||||
| 61 | Kim JY, 2010 [45] | Korea | Local | 60 | M | Overweight/obese: 9.5 ± 1.22 | Overweight/obese: 28.9 ± 0.20 | Plasma-Serum | UPLC-Q-TOF MS- Gas chromatography (GC) | Case/control | 8 |
| Normal weight: 39.6 ± 1.24 | Normal weight: 20.9 ± 0.14 | ||||||||||
| 62 | Pietiläinen KH, 2007 [76] | Finland | National | 14 |
F & M |
Pairs discordant for weight: (Non- | Serum | UPLC/MS-MS/MS | Cross sectional | 5 | |
| Obese co-twin = 25.4, Obese co-twin = 30.4) | |||||||||||
| 63 | [50] [50] | Lausanne, Switzerland | Local | 102 |
F & M |
Obese: > 25 | Plasma -Urine | NMR spectroscopy | Case/control | 5 | |
| Normal weight: < 21 | |||||||||||
| Under 18 years | |||||||||||
| ID | Author, Y | Country | Geographical Expansion national/subnational/international | Sample Size | Sex | Age | BMI | Sample | Method | Design | Score |
| 64 | Rauschert.S, 2017 [80] | Australia | National | 2900 |
F & M |
1, 2, 3, 5, 8, 10, 14, 17, and 20 | ND | Plasma EDTA samples | LC-MS/MS | Cohort | 7 |
| 65 | Cho K, 2016 [19] | Korea | Local | 200 |
F & M |
Control: 13.83 ± 0.43 Case: 13.84 ± 0.52 |
obese: BMI ≥ 30 | Urine | Untargeted metabolomic high performance liquid chromatography (LC)-quadrupole time-of-flight mass spectrometry (MS) and targeted metabolomic LC-MS/MS and flow injection analysis-MS/MS systems | Case/Control | 8 |
| 66 | Lee S H, 2016 [55] | South Korea | Local | 112 | F | 5 to 16 | Overweight (95th percentile > BMI 85th percentile) | Serum | LC–MS | Case/Control | 8 |
| Obesity (BMI 95th percentile) | |||||||||||
| Normal (85th percentile < BMI) | |||||||||||
| 67 | Gawlik,A, 2016 [30] | Silesia. | National | 87 |
F & M |
8.5–18.0 | >97 | Blood sample | GC-MS | Cohort | 8 |
| 68 | Butte NF, 2015 [15] | Spain | National | 803 |
F & M |
4–19 | ≥95 | Plasma samples | ultra-HPLC–tandem mass spectrometry | Cross sectional | |
| 69 | Perng W, 2014 [74] | U.S | International | 1116 |
F & M |
7.7 | ≥ 95 | Plasma samples | mass spectrometry | Cohort | 9 |
| 70 | Vitkin E, 2014 [95] | Israel | International | 394 |
F & M |
Obese = 4–17 | Normal = 13–29 Obese = 19–42 | Urine & blood | GC/MS | Case/Control | 8 |
| 71 | McCormack SE, 2013 [59] | USA | Subnational | 103 |
F & M |
8–18 |
Cross-sectional Cohort: 24.9 ± 7.4 Longitudinal Cohort baseline: 26.0 ± 7.1 Longitudinal Cohort 18 months: 27.9 ± 7.6 |
Plasma | LC-MS/MS | Cohort | 9 |
| 72 | [96] [96] | Germany | National | 120 |
F & M |
6 ≤ Age ≤ 15 | Normal: 17.2 ± 2.1 | Serum | LC-MS/MS | Case/Control | 9 |
| Obese: 27.7 ± 4.0 | |||||||||||
| 73 | Michaliszyn SF, 2012 [62] | Pennsylvania | Local | 139 |
F & M |
Obese: (normoglycemic 13.2 ± 0.2, dysglycemic 14.1 ± 0.3) Normal weight: 13.0 ± 0.2 |
Obese: (normoglycemic 32.5 ± 0.9, dysglycemic 35.5 ± 1.0) Normal weight: 18.9 ± 0.3 |
Plasma | Tandem mass spectrometry | Case/Control | 8 |
| 74 | Mihalik SJ, 2011 [62] | Pennsylvania | Local | 103 |
F & M |
Obese: 13.4 ± 0.23 | Overweight/obese: 34.6 ± 0.7 | Plasma | MS/MS | Case/Control | 8 |
| Normal weight: 13.0 ± 0.23 | Normal weight: 19.0 ± 0.3 | ||||||||||
| Normal weight: 13.0 ± 0.23 | Normal weight: 19.0 ± 0.3 | ||||||||||
Among included papers, 7 articles were assessed metabolic syndrome correlation with metabolites alterations. The 13 articles conducted for cases with the age range under 18 years old. The biological samples were applied for metabolomics analysis comprised of serum, plasma, urine, serum of venous cord blood, adipose tissue, cord-blood, placenta tissue, and exhaled breath condensate (EBC) samples. The most common samples between studies were serum and plasma which applied in 25 and 29 studies, respectively. The EBC, placenta, and adipose tissue, each one was used only in 1 study. In addition, 2 studies were used cord blood. The main experimental methods performing for metabolites identification were quantification include gas chromatography mass spectrometry (GC–MS), nuclear magnetic resonance (NMR), tandem mass spectrometry (MS/MS), liquid chromatography–MS (LC-MS), liquid chromatography-MS/MS (LC-MS/MS), the ultra-high performance liquid chromatography-quadrupole time-of-flight mass spectrometry (UHPLC/Q-TOF-MS), gas chromatography time-of-flight mass spectrometry (GC-TOF-MS), UPLC coupled to triple quadruple mass spectrometer (UPLC -TQ/MS), liquid chromatography coupled with flow-injection mass spectrometry (FIA-MS), Nexera X2 U- High-performance liquid chromatography (HPLC), Exactive plus orbitrap mass spectrometry, capillary electrophoresis– mass spectrometer (CE-MS), Beckman-Coulter clinical analyzer, Ultra performance liquid chromatography - mass spectrometer (UPLC-MS), UPLC-MS/MS, flow injection analysis electrospray ionization- mass spectrometer (FIA–ESI-MS), and FIA–MS. The most prevalent strategy was GC–MS used in 15 studies. In terms of specific metabolic targeting, 31 studies were just based on targeted metabolomics, 5 studies used both targeted and untargeted metabolomics profiling and 27 papers were reported to apply only untargeted metabolomics approaches. According to the information presented in the Table 1, most studies, approximately one-fourth of studies were conducted in the USA, followed by China and Korea.
Amino acids and related metabolites
Amino acids are associated with the process of obesity. In this section, a number of obesity-related amino acids are mentioned and have been reported in Supplementary Materials.
Body mass index & obesity
According to recent studies, a large number of amino acids have a close correlation with obesity. The most prominent category that is referred to in most studies is branched-chain amino acids (BCAAs) which include leucine, isoleucine, and valine. Since 2011, among the metabolomics studies on obesity, 18 studies have documented the direct relationship between leucine and isoleucine with obesity. Also, 22 studies highlight Valine’s importance among obesity biomarkers. Considering the importance of these amino acids, the approach of one of the studies examines the effect of these amino acids on the process of obesity. In all of these studies, it has been observed that as a result of obesity, the amount of these amino acids increases. Also, several studies have examined the possibility of tyrosine candidating as a biomarker for obesity. Among the papers under review in this report, 16 studies explicitly referred to increased tyrosine levels due to obesity. Only Badoud et al. investigated more specifically that the subjects were divided into 3 groups included metabolically healthy obese, metabolically unhealthy obese, and lean. The comparisons were done in three categories contains: metabolically healthy obese/ lean, metabolically unhealthy obese/ lean, and metabolically unhealthy obese/ metabolically healthy obese. The results show that the amount of tyrosine increased in the first and second comparisons, but in the last comparison, a decrease in the amount of tyrosine was observed. Another important amino acid is phenylalanine. Among the papers reviewed, 19 studies have examined the effect of phenylalanine on obesity, that 12 studies have indicated an increase in its amount due to obesity. Badoud et al. noticed a decrease in the amount of phenylalanine in the study of metabolically unhealthy obese subjects compared to metabolically healthy obese subjects. Also, Desert et al. performed four types of studies in three different groups, which in all studies reduced the amount of phenylalanine due to obesity. Another amino acid that has been studied is glycine, which there are 15 studies about it. Every 15 studies emphasize the reduction of glycine levels in obese subjects compared to healthy control subjects. In the case of glutamic acid, 13 studies have been done. In all cases, there was a direct relationship between obesity and increased glutamic acid levels. Also, Lysine has a relative importance in obesity and 12 studies have examined its effect on obesity. There are 9 reports that indicate an increase in the amount of lysine due to obesity. One study done by Desert et al. in comparing two overweight and normal weight groups shows that there is no difference in the amount of lysine between groups. Also, there are two reports that show a decrease in the Lysine level in obese subjects published by Fattuoni et al. and Palmnäs m, et al. Eventually Badoud et al., observed a decreased lysine level in metabolically unhealthy obese subjects rather than metabolically healthy obese subjects. The last amino acid that has a medium amount of reports is Alanine. There are 11 reports about the amount of alanine in obese subjects, in which all studies have indicated elevated levels of alanine due to obesity.
It seems that along with amino acids, their derivatives also play a role in the obesity process. Among the reviewed articles, the most frequent and most important derivatives studied are creatinine, creatine, kynurenine, urea, citrulline, ornithine, hyppurate, and serotonin. There are various reports on amino acid derivatives. For example, in the case of creatinine, a number of observations indicate an increase in its amount with obesity, including Yin, Desert, Valcarcel, and their colleague’s studies. Kim et al. and Valcarcel et al. in another examined group, referred to the decrease in its value. Also, in the case of creatine, there is a report that indicates a reduction in creatine level in obese subjects that this study was conducted by Schlecht et al. Another result from Ho et al. has been reported that which shows an increase in the amount of creatine associated with obesity. Desert et al. during four studies found that creatine levels increased in obese subjects, but in overweight individuals, its amount was not significantly different compared with healthy subjects. In a study on Kynurenine, Zhao, Ho, Chen and their colleagues reported an increased level of in a relationship with increased BMI. Sandler et al. and Valcarcel et al. studied urea and in association with obesity, each of them saw an increase in the amount of urea in a cohort and its reduction in another cohort. Two other important derivatives are citrulline and ornithine, which Isherwood, Okekunle, Kim, Ho, Xie and their colleagues have studied in this regard, that suggesting a decrease in citrulline level and decreased level of ornithine in obese subjects. The last metabolite is serotonin, which one report by Kim et al. indicates a negative association with obesity.
On the other hand, a number of studies have investigated the association of metabolites with fat mass, metabolic syndrome, and waist circumference.
Fat mass
In the context of the association of amino acids with fat mass, studies have been conducted by Murphy, Menni, Jourdan and their colleagues. The results indicate a direct correlation between the increase in the amount of BCAAs, tyrosine, phenylalanine, glutamic acid, and alanine by the increase in fat mass. Only one amino acid decreases with increasing fat mass, which is glycine.
Metabolic syndrome
Some studies have been conducted on metabolic syndrome. In this way, a comparison has been made between the metabolites profile of healthy controls and metabolic syndrome subjects. The results of these studies were reported by Okeknule, Allam-ndoul, Zhong and their colleagues. As a result of these reports, BCAAs, tyrosine, phenylalanine, glutamic acid, lysine and alanine increase in metabolic syndrome subjects compared to healthy controls. Only glycine decreased in obese subjects with metabolic syndrome compared to lean subjects.
Waist circumference
Waist circumference is one of the obesity indicators, which investigating its association with metabolites can be helpful to discover obesity biomarkers. Ho, Schlecht, Zhao and their colleagues have studied in this regard. As a general result, Ho et al. pointed to the direct correlation between the waist circumference and the amount of BCAAs, tyrosine, phenylalanine, and alanine and reverse relationship with glycine. Also, Schlecht et al. found an increase in the amount of alanine and decreased level of phenylalanine due to increased waist circumference. Zhao et al. only achieved an increase in the amount of glutamic acid associated with waist circumference. Finally, Rauschert et al. observations indicate increased levels of leucine, tyrosine, and phenylalanine due to waist circumference in subjects that are under 18 years.
Under 18 years
Studies show that adolescence can affect biomarkers of obesity. For this reason, some studies have examined obesity under the age of 18 years. In this field Rauscher, Cho, Butte, Perng, McCormack, Michaliszyn and their colleagues have reported their results. The results indicate the stability of the biomarkers mentioned in the previous section under 18 years. There is only a report of Cho et al. that indicates a decrease in the amount of isoleucine in obese subjects who are under the age of 18 years.
Lipid derivatives
Body mass index & obesity
In terms of lipid derivatives, Isherwood CM, Tulipani S, Rauschert S, Kim Y J, Hellmuth C, and Dunn WB and their colleagues have revealed that decreased levels of LPCs promote obesity in adults [24, 33, 42, 46, 79, 92]. Kim Y J showed that among different LPCs which decrees in obese adults, LPC C16:0 increases in obese people [46]. Pietiläinen KH, et al. in 2007 were the only group found the higher concentrations of LPCs in obesity condition [76]. Hellmuth C, et al. showed that elevated levels of acyl-LPCs in cord blood are highly associated with birth weight [34]. Shokry et al. in 2019, has been studied the impact of maternal prepregnancy BMI on both maternal and cord blood metabolic profiles that reported results shown decreased levels of LPCs. In line with that, Bagheri, Wang and their colleagues have been shown decreased LPC metabolites in obese adults. Kochhar S, et al. were the first group to suggest that a lower level of choline is related to obesity [50]. But Ho JE, et al. reported that choline concentration in obesity and elevated WC is higher than normal weight and the glycerol-phosphocholine level is decreased in obese adults [36]. Ahmad MS, et al. indicated that phosphorylcholine is decreased in obese adults [3]. After that, Schlecht I, et al., demonstrated that a lower concentration of choline was detected in obese individuals [35]. Chen H, and Dugas L, announced that decreased amount of glycerophosphocholine was seen in obese adults [18, 23]. Among three populations investigated in Dugas L, et al. study South African obese adults showed a lower level of glycerophosphocholine and Ghanaian people vice versa [23]. There is controversial evidence about the role of diacylphosphatidylcholines (PCaa) and phosphatidylcholines (PCae) in obesity progression. Rauschert S, and Jourdan C, claimed the positive relation between PC aa and obesity [44, 79] while Isherwood CM, et al. showed the negative relation [42]. and Cho K, 2016 [19] revealed that some PC aa exhibit positive and some others show a negative correlation with obesity in adults, and obesity in childhood, respectively [19, 36, 46]. Cho K, et al. claimed some PC ae showed positive and some others negative association with obesity in adolescents [19]. Bagheri M, et al. showed a positive association of obesity with PCaa and a negative association with PCae [11]. Similarly, Shokry E, et al. in their cohort study on pregnant mothers demonstrated the elevated levels of PCaa in overweight/obese mothers. As well they reported a decrease in PCae levels [88]. On the one hand, Rauschert S, and Hellmuth C, found a positive relation between SMs and adult obesity [33, 79]. On the other hand, Dunn WB, and Kim Y J, demonstrate lower concentrations of SMs in adult obese and Cho K, et al. reported the same results in obese youth [19, 24, 46]. ACs with different acyl chain lengths were addressed by Sandler V, Isherwood CM, and Allam-Ndoul B, and showed a positive association with adult obesity and higher BMI [4, 42, 84]. NEFA was another lipid derivative known as an obesity contributor. Rauschert S, and Hellmuth C, reported this association [33, 79].
Fat mass and waist circumference
Rauschert S and Ho JE in 2016 and also Jourdan C in 2012, showed that lower levels of LPCs are positively related to higher WC and fat mass, respectively [36, 44, 79]. Schlecht I, et al., shown that a lower concentration of choline was detected in persons with higher fat mass and WC [85]. Rauschert S, and Jourdan C, demonstrated positive relation between PC aa and increased WC [79] and fat mass [44], while Isherwood CM, et al. showed a negative relation [42]. Ho JE, 2016 revealed a negative correlation with higher WC in adults [36]. Furthermore, Jourdan C, et al. reported that decreased concentrations of PCae is associated with higher fat mass [28]. Rauschert S, et al. revealed a high level of PC ae in people with higher WC [79]. Rauschert S, and Ho JE, showed an elevated level of SMs in people with higher WC [36, 79]. Jourdan C, et al. demonstrated that an increased level of C5 is positively related to higher fat mass leading to obesity [44]. Rauschert S, showed an elevated concentration of NEFA is positively correlated with higher WC [79].
Metabolic syndrome
Allam-Ndoul B, showed that short-chain ACs such as C0, C3, and C5 are correlated with obesity and MetS. Moreover, they showed that the levels of some long-chain ACs like C36, C40, and C42 are inversely related to obesity and MetS [4].
Under 18 years
Despite a positive correlation between NEFA and obesity, Hellmuth C, showed that higher cord blood levels of NEFA C22:6 and NEFA C20:5 were associated with lower birth weight [34]. Furthermore, Wahl S, et al. demonstrated lower level of PC ae in obese children [44, 96].
Carbohydrate metabolism derivatives
Glucose and glycerol are positively correlated with obesity. In terms of lactate, conflicting evidence hinders the clarification of exact effect of this metabolite on obesity. Lower concentrations of acetate and predominantly citrate have been suggested in the obese population. Pyruvate is increased in obesity state.
Body mass index & obesity
There is a consensus that glucose is positively correlated with obesity. Ahmad MS, Lin Z, and Valcárcel B, revealed that glucose level is increased in obese adults [3, 57, 93]. Additionally, Fattuoni C, et al. displayed higher glucose-6-phosphate levels in obese adults [26]. Six articles evaluated the glycerol concentration in obese adults and indicated the positive relationship between obesity and glycerol levels [26, 36, 84, 93, 104, 105]. Because the lactate assessment displayed conflicting results we are not able to reach a consensus reflecting the precise effect of lactate on obesity. On the one hand, Schlecht I, Kochhar S, and Paris D showed obesity is inversely associated with lactate levels in obese adults [50, 69, 85]. Schlecht I, et al. also reported the same result related to higher WC [85]. On the other hand, Yin X, Ho JE, Desert R, Valcárcel B, and Xie G, showed increased concentrations of lactate in obese adults [22, 36, 93, 104, 105]. Ahmad MS, Paris D, Valcárcel B, and Dunn WB, have reported a lower concentration of acetate in obese adults [3, 24, 69, 93]. Yin X, Valcárcel B, Xie G, and Butte NF, demonstrated that pyruvate as the final product of glycolysis was increased in obese adults [93, 104, 105]. Citrate as one of the TCA cycle intermediate is mainly reported to be decreased in obese adults by Ahmad MS, and Valcárcel B [3, 93]. Menni C, found the negative association between citrate level and fat mass in obese adults [60]. Kochhar S, showed lower and higher citrate concentration in plasma and urine of obese adults, respectively [50]. Despite the majority of studies, Desert R, claimed an increased level of citrate in obese adults [22].
Fat mass and waist circumference
Menni C, et al. reported the positive relationship between adult fat mass and glucose concentration [60]. Ho JE, et al. also reported these same relations between elevated WC and glycerol [36]. Consistent with these results, Menni C, and Ho JE, claimed the lactate positive relation with fat mass and WC, respectively [36, 60].
Under 18 years
Butte NF, demonstrated that pyruvate as the final product of glycolysis was increased in obese children. They also reported a decrease in citrate in obese children [15].
Nucleic acids metabolism derivative
Higher concentration of urate has been measured in obese people highlighting that increased levels of urate may contribute to obesity. Yin X, Ho JE, and Dunn WB indicated that urate as one of the nucleic acids metabolism intermediates showed an increased level in obese adults [24, 36, 105]. Moreover, Menni C, et al. announced the positive correlation between urate concentration and fat mass in obese adults [60].
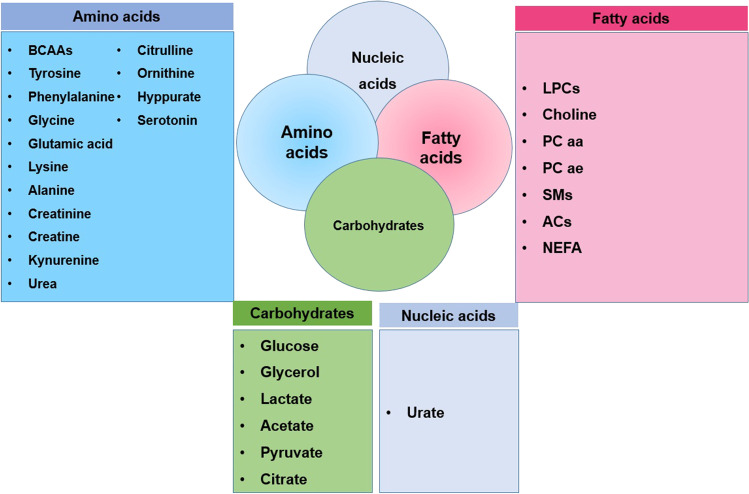
The most common metabolites
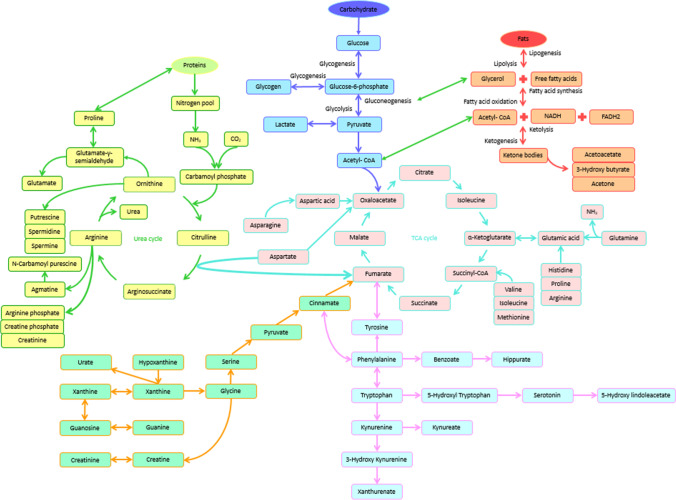
The most common metabolites based on the systematical review conducted in present study are provided in Fig. 2. More detailed information of different metabolites is available in Supplementary Materials (Fig. 3).
Fig. 2.
The most common metabolites associated with obesity
Fig. 3.
consists of major biochemical cycles and shows the relevance of important metabolites, that most of them are effective in obesity. Biochemical cycles are closely related with each other. So that the product of each cycle is used as a primary material or auxiliary agent in another cycle. Thus, the overall biochemical cycle is formed for the metabolism of the living organism. Certainly, fluctuations in the metabolites due to disease or genetic defects are affecting this pathway and cause consecutive fluctuations in the amount of metabolites. Therefore, tracking these fluctuations can act as a good biomarker to diagnose diseases and even to develop a therapeutic method for them. Here the focus of the study is on obesity and a review of the overall metabolic cycle based on the metabolites that are effective in obesity. Based on the studies conducted in this article, metabolites that are effective in obesity have been identified. Also, most obesity biomarkers are present in the overall metabolic cycle. (Figure 3) Therefore, it is possible to follow the process of obesity and develop a therapeutic method for it. Meanwhile, there are a number of important biomarkers in obesity that include amino acids such as phenylalanine, tyrosine, lysine, glutamic acid, alanine and BCAA (branched chain amino acids), polyamines that most notably is putrescine and ketone bodies including acetoacetate and 3-hydroxy butyrate
Discussion
Amino acids and related metabolites
Metabolites are intermediates and products derived from the metabolism of living cells. Amino acids are important and essential metabolites in the body that play an important role in the metabolism of the body, which includes three key roles: the necessary substrate for protein synthesis, providing nitrogen for the synthesis of other nitrogenous compounds, Catabolization as a fuel, and energy source that can be converted to precursors for the production of carbohydrates and lipids. Our studies showed that some of the amino acids are associated with metabolic diseases such as obesity. The overall result of the studies was an increase in the amount of leucine, isoleucine, valine, tyrosine, phenylalanine, glutamic acid, lysine and alanine, and a decreased glycine level in obese subjects were observed. In general, it may be possible to associate increased levels of some amino acids with low expression of the LAT1 protein in obese subjects, which is responsible for the transport of large natural amino acids, including BCAAs, tyrosine and phenylalanine [10] BCAAs, like other amino acids, play important roles in the body. Regarding their association with obesity, there are reports that the mitochondrial activity of branched-chain amino acid aminotransferase and branched chain-α- keto acid dehydrogenase enzymes are reduced in adipose tissue of obese subjects. These are key enzymes of the BCAAs catabolic pathway, which reduction in their activity leads to an increased level of BCAAs (Baogang. X. 2012).
Regarding changes in the number of aromatic amino acids, including phenylalanine and tyrosine, there are many assumptions that have not been fully understood. Their changes can be explained by several reasons. One of the reasons is that these amino acids compete with the increased level of BCAAs for absorption by tissues, which increases their circulating amounts in blood. Another hypothesis is liver dysfunction due to metabolic disorders, which leads to a decrease in the metabolism of phenylalanine and tyrosine, and ultimately increases their levels in the blood (Diane M. Libert.2018).
Glutamic acid is a basic substance for energy metabolism associated with metabolic diseases. High levels of glutamic acid in obese individuals are due to the lower absorption by the TCA cycle. It seems that mitochondrial TCA dysfunction is associated with an increase in glutamic acid in obese subjects. In addition, glutamic acid may also be triggered by glucagon release from alpha-pancreatic cells exacerbating metabolic diseases. As a result, the increase in pyruvate to alanine transamination increases the concentration of alanine, as well as an accelerator of the gluconeogenesis process and increases the amount of glucose in the blood [65].
Glycine has a protective effect that can leads to a reduction in the mitochondrial Acetyl-CoA through the formation of Acetyl-glycine in the kidney. This process stimulates the oxidation of fatty acids in mitochondria that in obese people, this pathway is disturbed. In addition, in the treatment of obesity, glycine supplementation is an effective way to accelerate fat loss and prevent muscle loss in obese people (Guevara-cruz. M.2018).
Finally, in this review, we have found that most of the amino acids and their derivatives have closely interlinked with obesity, which each of them has the ability to be used as a biomarker for obesity.
Lipid derivatives
LPCs as important lipidic intermediates which are mainly decreased in obese populations are formed by the lipoprotein-associated enzyme called lecithin cholesterol acyltransferase (LCAT) responsible for esterification of cholesterol [82]. LCAT acts by cleaving fatty acids from PCs and transferring them onto cholesterol [43, 82]. The level of LCAT is inversely correlated with SM concentration [89]. As mentioned before, in obese people with increased BMI, SM species are elevated and leading to reduced activity of LCAT. Therefore, increased concentrations of PCs are potentially due to a decrease in LCAT activity. Consequently, the accumulation of PCs has occurred with no esterification process of cholesterol [79].
Pietiläinen KH, et al. studied obesity in twins suggested that regardless of genetic material metabolite alterations may contribute to atherosclerosis and diabetes [76]. Contrary to other studies, they claimed that increased concentrations of LPCs may be related to oxidative stress and also endothelial dysfunction which was revealed in preclinical studies by Galili O, et al. [28, 76].
Jourdan C, et al. recommended that the inverse correlation of very long chain PC spices and positive relation of PC aa C38:3 with fat mass is potently due to more functional activity of enzymes responsible for very-long chain fatty acids oxidation. They also found that chain elongation and fatty acid desaturation enzymes may be involved in this process [44].
Higher concentrations of SM species which were correlated with obesity and high WC are potentially a consequence of elevated SMs biosynthesis. Ceramide plays a role in SM species biogenesis and ceramide-choline phosphotransferase catalyzes the binding of phosphocholine to a ceramide molecule [29]. Sphingosine as the precursor of SM species is formed by the action of ceramidase in order to produce an enhanced level of SM species [14].
Stearoyl-CoA desaturase-1 (SCD-1) seems as one of the key players of lipogenesis instead of lipidic β-oxidation leading to fetal fat accumulation and higher risks for increased birth weight and following obesity in childhood [40, 51]. ACs are major components for transportation of lipids and proteins catabolites into mitochondria resulting in breaking down of these molecules through β-oxidation reactions and energy supply [18].
Increased levels of ACs in adults and adolescents have been approved previously. This process is aided by carnitine palmitoyltransferase 1 (CPT1), a mitochondrial enzyme, that catalyzes the production of ACs via acyl transfer to l-carnitine which is required for transportation of acyl group into mitochondria intended for β-oxidation process [37].
Increased levels of short length ACs also could be attributed to greater availability of NEFA or decreased oxidation of NEFA [61]. AC-C3, and LPCs (C18:1 and C18:2) are considered as potential biomarkers of obesity with T2DM [42].
Although it has been reported that NEFA in obese adults is higher than normal, Hellmuth C, et al. found that lower levels of cord blood NEFA is associated with reduced lipolytic activity and elevated fetal NEFA uptake in adipose tissue [34, 49]. During embryonic development, adipose tissue expansion has occurred and an enhanced level of NEFA is uptook by fatty tissue is led to reduced concentration of cord blood NEFA [34]. During gestation, an increase in BMI is commonly due to fat deposits, mainly NEFA, in adipose tissue prior to pregnancy [32].
It is not worthy that the size and composition of side chains of various spices of PC, LPC, SM, and AC is a pivotal element that imposes different and somehow opposite effects on human health. Because of a wide range of variability among lipid derivatives, it is difficult to interpret their exact involvement in obesity or some other metabolic disorders.
Carbohydrate metabolism derivatives
Glycerol is introduced as a well-known lipid metabolism component involved in supplying energy to the cells. The elevated level of glycerol in obesity conditions may come from higher concentrations of fatty acids. In the case of obese placentas of obese pregnancies, higher levels of placental fatty acids led to enhanced fatty acids uptake by the fetus [16].
Preclinical and clinical evidence revealed that citrate as a TCA cycle metabolite is inversely related to obesity [25, 48]. It has been described that rats with high fat diet which developed diabetes, the TCA cycle intermediates such as lactate and citrate have decreased. Additionally, the higher activity of β-oxidation reactions led to a reduced level of TCA cycle intermediates in animals suffering from insulin resistance [52].
Lactate could reflect the cell oxidative capacity and its enhanced concentration is generally sound as a biomarker for some metabolic disorders like type 2 diabetes [6, 21]. Lactate plays a role as a precursor of gluconeogenesis and increased concentration of lactate may be owning to glucose and glycogen biosynthesis problems [22].
Conclusion
In summary, this systematic review summarized available evidence for the relevance between metabolomics and metabolite profile of obesity. As the first systematic review in this area, the present study will be a precious source for both researchers and clinicians to transparent the informational gaps in the management of obesity. Although the wide range of studies has been covered by this systematic review regardless of the age, history, language, and type of the study, further studies are needed to compare the application of emerging methods in the treatment of obesity and related disorders. Eventually, in the future studies, more reliable and quantitative results will be achieved through a meta-analysis of data.
Supplementary Information
(RAR 529 kb)
A
- AC-C0
Acylcarnitine-C0
- AC-C2
Acylcarnitine-C2
- AC-C3
Acylcarnitine-C3
- AC-C4
Acylcarnitine-C4
- AC C4-OH
Acylcarnitine C4-OH
- AC C5
Acylcarnitine C5
- AC C8
Acylcarnitine C8
- AC C8:1
Acylcarnitine C8:1
- AC C10
Acylcarnitine C10
- AC C10:1
Acylcarnitine C10:1
- AC C10:2
Acylcarnitine C10:2
- AC C10:3
Acylcarnitine C10:3
- AC C12:1
Acylcarnitine C12:1
- AC-C14:1
Acylcarnitine-C14:1
- AC-C16
Acylcarnitine-C16
- AC C16-OH/C14-DC
Acylcarnitine C16-OH/C14-DC
- AC C16:1
Acylcarnitine C16:1
- AC-C18
Acylcarnitine-C18
- AC C18:1
Acylcarnitine C18:1
- AC C18:1-OH/C16:1-DC
Acylcarnitine C18:1-OH/C16:1-DC
- ADMA
Asymmetric dimethylarginine
- AHB
α-hydroxybutyrate
- AKB
2-AMINO-3-KETOBUTYRIC ACID
- alpha-AAA
alpha-amino adipic acid
- Arg
Arginine
- Asn
Asparagine
B
- BHBA
Beta-Hydroxybutyric acid
C
- C0
Carnitine (free)
- C3
Propionylcarnitine
- C14:1
Tetradecadienoylcarnitine (C14:1)
- C14:1-OH
3-Hydroxymyristoleylcarnitine
- C14:2
Tetradecadienoylcarnitine (C14:2)
- C16:0
Hexadecanoic acid
- C16:1
Palmitoleic acid
- C18:0 LPE
C18:0 lysophosphatidyl-ethanolamine
- C18:1
Oleic acid
- C18:1 LPC
C18:1 lysophosphatidylcholine
- C18:1 LPE
C18:1 lysophosphatidyl-ethanolamine
- C18:2 LPC
C18:2 lysophosphatidylcholine
- C20:3 CE
C20:3 cholesterol ester
- C20:5 CE
C20:5 cholesterol ester
- C22:1
Erucic acid
- C22:2
c -13,16-Docosadienoic acid
- C22:5n-6
Dpan-6
- C22:6 CE
C22:6 cholesterol esters
- C24:0
Tetracosanoic acid
- C24:1
Nervonic acid
- C30:0 DAG
C30:0 diacylglycerol
- C32:0 DAG
C32:0 diacylglycerol
- C32:1
Dotriacontenylic acid
- C32:1 DAG
C32:1 diacylglycerol
- C32:2 DAG
C32:2 diacylglycerol
- C34:0 DAG
C34:0 diacylglycerol
- C34:1
Tetratriacontenylic acid
- C34:1 DAG
C34:1 diacylglycerol
- C34:1 PC plasmalogen A
C34:1 Phosphatidylcholine plasmalogen A
- C34:2
Tetratriacontadienoic acid
- C34:2 DAG
C34:2 diacylglycerol
- C34:3
Acyl-akyl-phosphatidylcholine
- C34:3 DAG
C34:3 diacylglycerol
- C34:4 PC
C34:4 Phosphatidylcholine
- C36:0
Hexatriacontanoic acid
- C36:0 DAG
C36:0 diacylglycerol
- C36:1 DAG
C36:1 diacylglycerol
- C36:1 PC plasmalogen
C36:1 Phosphatidylcholine plasmalogen
- C36:2
Hexatriacontadienoic acid
- C36:2 DAG
C36:2 diacylglycerol
- C36:2 PC plasmalogen
C36:2 Phosphatidylcholine plasmalogen
- C36:3 DAG
C36:3 diacylglycerol
- C36:3 PC plasmalogen
C36:3 Phosphatidylcholine plasmalogen
- C36:4 DAG
C36:4 diacylglycerol
- C38:0
Octatriactanoic acid
- C38:3 PC
C38:3 Phosphatidylcholine
- C38:4 DAG
C38:4 diacylglycerol
- C38:5 DAG
C38:5 diacylglycerol
- C38:6 PC
C38:6 Phosphatidylcholine
- C38:7 PE plasmalogen
C38:7 Phosphatidylethanolamine plasmalogen
- C40:6 PE
C40:6 Phosphatidylethanolamine
- C40:9 PC
C40:9 Phosphatidylcholine
- C46:2 TAG
C46:2 triacylglycerol
- C46:3 TAG
C46:3 triacylglycerol
- C46:4 TAG
C46:4 triacylglycerol
- C48:1 TAG
C48:1 triacylglycerol
- C48:2 TAG
C48:2 triacylglycerol
- C48:3 TAG
C48:3 triacylglycerol
- C48:4 TAG
C48:4 triacylglycerol
- C50:0 TAG
C50:0 triacylglycerol
- C50:1 TAG
C50:1 triacylglycerol
- C50:2 TAG
C50:2 triacylglycerol
- C50:3 TAG
C50:3 triacylglycerol
- C50:4 TAG
C50:4 triacylglycerol
- C50:5 TAG
C50:5 triacylglycerol
- C50:6 TAG
C50:6 triacylglycerol
- C52:0 TAG
C52:0 triacylglycerol
- C52:1 TAG
C52:1 triacylglycerol
- C52:2 TAG
C52:2 triacylglycerol
- C52:3 TAG
C52:3 triacylglycerol
- C52:4 TAG
C52:4 triacylglycerol
- C52:5 TAG
C52:5 triacylglycerol
- C52:6 TAG
C52:6 triacylglycerol
- C52:7 TAG
C52:7 triacylglycerol
- C54:1 TAG
C54:1 triacylglycerol
- C54:2 TAG
C54:2 triacylglycerol
- C54:6 TAG
C54:6 triacylglycerol
- C54:7 TAG
C54:7 triacylglycerol
- C54:8 TAG
C54:8 triacylglycerol
- C54:9 TAG
C54:9 triacylglycerol
- C56:5 TAG
C56:5 triacylglycerol
- C56:6 TAG
C56:6 triacylglycerol
- C56:7 TAG
C56:7 triacylglycerol
- C56:8 TAG
C56:8 triacylglycerol
- C56:9 TAG
C56:9 triacylglycerol
- C56:10 TAG
C56:10 triacylglycerol
- C58:6 TAG
C58:6 triacylglycerol
- C58:7 TAG
C58:7 triacylglycerol
- C58:8 TAG
C58:8 triacylglycerol
- C58:9 TAG
C58:9 triacylglycerol
- C58:10 TAG
C58:10 triacylglycerol
- C58:11 TAG
C58:11 triacylglycerol
- CE
Cholesterol ester
- CE(20:3)
cholesterol ester (20:3)
- CE(22:5)
cholesterol ester (22:5)
- CE(22:6)
cholesterol ester (22:6)
- Cer(d18:0/23:0)
ceramides(d18:0/23:0)
- Cer(d18:1/18:0)
ceramides(d18:1/18:0)
D
- DG(44:5)
Diacylglycerol (44:5)
- DHEA-S
Dehydroepiandrosterone sulfate
G
- Glu
Glutamic acid
- Gly
Glycine
H
- HDL
High-density lipoprotein
- His
Histidine
L
- Leu
Leucine
- LPA 16:0
[(2R)-2-(hexadecanoyloxy)-3-hydroxypropoxy]phosphonic acid
- LPC
Lysophosphatidylcholines
- LPCa C14:0
lysoPhosphatidylcholine a C14:0
- LPCa C16:0
lysoPhosphatidylcholine a C16:0
- LPC a c16:0 / LPCa C20:3
lysophophatidylcholine
- LPC a c16:0 / LPCa C20:4
lysophophatidylcholine
- LPC a c16:0 / PC aa C32:0
lysophophatidylcholine
- LPC a c16:0 / PC aa C36:2
lysophophatidylcholine
- LPCa C16:1
lysoPhosphatidylcholine a C16:1
- LPC a c18:0/ LPCa C20:3
lysophophatidylcholine
- LPC a c18:0 / LPCa C20:4
lysophophatidylcholine
- LPC a c18:0 / PC aa C36:2
lysophophatidylcholine
- LPC a c18:0 / PC aa C36:1
lysophophatidylcholine
- LPC Ac18:1
lysophophatidylcholine
- LPC Ac18:2
lysophophatidylcholine
- LPCa C18:3
lysoPhosphatidylcholine a C18:3
- LPCa C20:3
lysoPhosphatidylcholine a C20:3
- LPCa C20:4
lysoPhosphatidylcholine a C20:4
- LPC Ac20:4
lysophophatidylcholine
- LPE
Lysophosphatidylethanolamines
- LysoPC(18:1)
lysoPhosphatidylcholine (18:1)
- LysoPC(18:2)
lysoPhosphatidylcholine (18:2)
- LysoPC(20:1)
lysoPhosphatidylcholine (20:1)
- lysoPC a C16:0
lysoPhosphatidylcholine acyl C16:0
- LysoPC a C17:0
Lysophosphatidylcholine a C17:0
- lysoPC a C17:0
lysoPhosphatidylcholine acyl C17:0
- LysoPC a C18:0
lysoPhosphatidylcholine a C18:0
- lysoPC a C18:0
lysoPhosphatidylcholine acyl C18:0
- lyso.PC.a.C18.1
lysoPhosphatidylcholine a C18:1
- lysoPC a C18:1
lysoPhosphatidylcholine acyl C18:1
- LysoPC a C18:2
lysoPhosphatidylcholine a C18:2
- lysoPC a C18:2
lysoPhosphatidylcholine acyl C18:2
- lyso.PC.a.C18.3
lysoPhosphatidylcholine a C18:3
- lysoPC a C20:4
lysoPhosphatidylcholine a C20:4
- lysoPC a C26:0
lysoPhosphatidylcholine acyl C26:0
- lyso.PC.e.C16.0
lysoPhosphatidylcholine a C16.0
- lyso.PC.e.C18.0
lysoPhosphatidylcholine a.C18.0
- LysoPE(22:4)
lysoPhosphatidylcholine (22:4)
- LysoPE a 18:0
Lysophosphatidylethanolamine(0:0/18:0)
- LysoPE a 18:1
Lysophosphatidylethanolamine(18:1/0:0)
- LysoPE a 18:2
Lysophosphatidylethanolamine(18:2)
N
- N-C18-1-Cer
N-(9Z-octadecenoyl)-ceramide; N-(oleoyl)-ceramide
- NEFA.12.1
non-esterified fatty acids
- NEFA.14.0
non-esterified fatty acids
- NEFA.14.1
non-esterified fatty acids
- NEFA.14.2
non-esterified fatty acids
- NEFA.14.4
non-esterified fatty acids
- NEFA 15:0
non-esterified fatty acids
- NEFA.16.0
non-esterified fatty acids
- NEFA.16.1
non-esterified fatty acids
- NEFA.16.2
non-esterified fatty acids
- NEFA.17.0
non-esterified fatty acids
- NEFA.17.1
non-esterified fatty acids
- NEFA 18:1
non-esterified fatty acids
- NEFA.18.2
non-esterified fatty acids
- NEFA.18.3
non-esterified fatty acids
- NEFA.18.4
non-esterified fatty acids
- NEFA.19.1
non-esterified fatty acids
- NEFA 20:1
non-esterified fatty acids
- NEFA.20.2
non-esterified fatty acids
- NEFA 20:3
non-esterified fatty acids
- NEFA 20:4
non-esterified fatty acids
- NEFA.20.5
non-esterified fatty acids
- NEFA 22:4
non-esterified fatty acids
- NEFA 22:5
non-esterified fatty acids
- NEFA C20:5
non-esterified fatty acids C20:5
- NEFA C22:6
non-esterified fatty acids C22:6
P
- PA(28:0)
Phosphtatidic acid (28:0)
- PC
Phosphatidylcholine
- PC(16:0/O-1:0)
Phosphatidylcholine(16:0/O-1:0)
- PC(16:0/O-16:0)
Phosphatidylcholine (16:0/O-16:0)
- PC(18:3/dm18:1)
Phosphatidylcholine(18:3/dm18:1)
- PC(19:3)
Phosphatidylcholine(19:3)
- PC(22:4/dm18:1)
Phosphatidylcholine(22:4/dm18:1)
- PC(35:2)
Phosphatidylcholine(35:2)
- PCA
2-Pyrrolidone-5-carboxylic acid
- PC aa C28:1
Phosphatidylcholine diacyl C28:1
- PC aa C30:2
Phosphatidylcholine diacyl C 30:2
- PC aa C32:0
Phosphatidylcholine diacyl C32:0
- PC aa C32:1
Phosphatidylcholine diacyl C32:1
- PC.aa.C32.3
Phosphatidylcholine diacyl C32.3
- PC aa C34:1
Phosphatidylcholine diacyl C34:1
- PC aa C34:2
Phosphatidylcholine diacyl C34:2
- PC aa C34:3
Phosphatidylcholine diacyl C34:3
- PC aa C34:4
Phosphatidylcholine diacyl C34:4
- PC.aa.C34.5
Phosphatidylcholine diacyl C34.5
- PC aa C36:0
Phosphatidylcholine diacyl C36:0
- PC aa C36:1
Phosphatidylcholine diacyl C36:1
- PC aa C36:2
Phosphatidylcholine diacyl C36:2
- PC aa C36:3
Phosphatidylcholine diacyl C36:3
- PC aa C36:4
Phosphatidylcholine diacyl C36:4
- PC aa C36:5
Phosphatidylcholine diacyl C36:5
- PC aa C36:6
Phosphatidylcholine diacyl C36:6
- PC aa C38:0
Phosphatidylcholine diacyl C38:0
- PC aa C38:1
Phosphatidylcholine diacyl C38:1
- PC.aa.C38.3
Phosphatidylcholine diacyl C38:3
- PC.aa.C38.4
Phosphatidylcholine diacyl C38:4
- PC aa C38:5
Phosphatidylcholine diacyl C38:5
- PC aa C38:6
Phosphatidylcholine diacyl C38:6
- PC aa C40:0
Phosphatidylcholine diacyl C40:0
- PC aa C40:1
Phosphatidylcholine diacyl C40:1
- PC aa C40:2
Phosphatidylcholine diacyl C40:2
- PC aa C40:3
Phosphatidylcholine diacyl C40:3
- PC.aa.C40.4
Phosphatidylcholine diacyl C40.4
- PC.aa.C40.5
Phosphatidylcholine diacyl C40:5
- PC aa C40:6
Phosphatidylcholine diacyl C40:6
- PC aa C42:0
Phosphatidylcholine diacyl C42:0
- PC aa C42:1
Phosphatidylcholine diacyl C42:1
- PC.aa.C42.2
Phosphatidylcholine diacyl C42.2
- PC aa C42:5
Phosphatidylcholine diacyl C42:5
- PC aa C42:6
Phosphatidylcholine diacyl C42:6
- PC.aa.C43.4
Phosphatidylcholine diacyl C43:4
- PC.aa.C44.12
Phosphatidylcholine diacyl C44.12
- PC ae C32:1
Phosphatidylcholine acyl-alkyl C32:1
- PC ae C32:2
Phosphatidylcholine acyl-alkyl C32:2
- PC ae C34:1
Phosphatidylcholine acyl-alkyl C34:1
- PC.ae.C34.2
Phosphatidylcholine acyl-alkyl C34.2
- PC ae C34:3
Phosphatidylcholine acyl-alkyl C34:3
- PC ae 36:0
Phosphatidylcholine acyl-alkyl 36:0
- PC ae 36:1
Phosphatidylcholine acyl-alkyl 36:1
- PC ae 36:2
Phosphatidylcholine acyl-alkyl C 36:2
- PC ae 36:3
Phosphatidylcholine acyl-alkyl C 36:3
- PC ae 36:4
Phosphatidylcholine acyl-alkyl36:4
- PC.ae.C36.5
Phosphatidylcholine acyl-alkyl C36.5
- PC ae C38:0
Phosphatidylcholine acyl-alkyl C38:0
- PC ae C38:1
Phosphatidylcholine acyl-alkyl C38:1
- PC ae C38:2
Phosphatidylcholine acyl-alkyl C38:2
- PC.ae.C38.3
Phosphatidylcholine acyl-alkyl C38.3
- PC ae C38:4
Phosphatidylcholine acyl-alkyl C38:4
- PC ae C38:5
Phosphatidylcholine acyl-alkyl C38:5
- PC ae C38:6
Phosphatidylcholine acyl-alkyl C44:4
- PC ae C40:1
Phosphatidylcholine acyl-alkyl C40:1
- PC ae C40:2
Phosphatidylcholine acyl-alkyl C40:2
- PC ae C40:3
Phosphatidylcholine acyl-alkyl C40:3
- PC ae C40:4
Phosphatidylcholine acyl-alkyl C40:4
- PC ae C40:5
Phosphatidylcholine acyl-alkyl C40:5
- PC ae C42:0
Phosphatidylcholine acyl-alkyl C42:0
- PC ae C42:1
Phosphatidylcholine acyl-alkyl C42:1
- PC ae C42:2
Phosphatidylcholine acyl-alkyl C42:2
- PC ae C42:3
Phosphatidylcholine acyl-alkyl C42:3
- PC ae C42:4
Phosphatidylcholine acyl-alkyl C42:4
- PC ae C42:5
Phosphatidylcholine acyl-alkyl C42:5
- PC ae C44:3
Phosphatidylcholine acyl-alkyl C44:3
- PC ae C44:4
Phosphatidylcholine acyl-alkyl C44:4
- PC ae C44:5
Phosphatidylcholine acyl-alkyl C44:5
- PC(O-10:0/O-8:0)
Phosphatidylcholine(O-10:0/O-8:0)
- PC(O-10:0/O-10:0)
Phosphatidylcholine(O-10:0/O-10:0)
- PC (O-10:0/O-12:0)
Phosphatidylcholine (O-10:0/O-12:0)
- PE(22:1/dm18:1)
Phosphatidylethanolamine(22:1/dm18:1)
- PE(22:4/dm18:0)
Phosphatidylethanolamine(22:4/dm18:0)
- PG(38:3)
Prostaglandin(38:3)
- Phe
Phenylalanine
- PS(24:0)
Phosphtatidylserines(24:0)
S
- SDMA
Symmetric dimethylarginine
- SFA
Saturated fatty acid
- SM
Sphingomyelin
- SM C16:0 or SM (d18:1/16:0)
n-(hexadecanoyl)-sphing-4-enine-1-phosphocholine
- SM C24:1
n-(hexadecanoyl)-sphing-4-enine-1-phosphocholine
- SM (d16:1/18:0)
N-(octadecanoyl)-hexadecasphing-4-enine-1-phosphocholine
- SM(d18:0/20:0)
Sphingomyelin(d18:0/20:0)
- SM(d18:1/16:0)
Sphingomyelin(d18:1/16:0)
- SM (d18:2/16:0)
N-(hexadecanoyl)-4E,14Z-sphingadienine-1-phosphocholine
- SM (d18:2/18:0)
N-(octadecanoyl)-4E,14Z-sphingadienine-1-phosphocholine
- SM (OH) C14:1
Hydroxysphingomyeline C14:1
- SM (OH) C16:1
HydroxySphingomyelin C16:1
- SM (OH) C22:1
N-[(13Z)-3-Hydroxydocos-13-enoyl]sphing-4-enine-1-phosphocholine
- SM (OH) C22:2
HydroxySphingomyelin C22:2
- SM (OH) C24:1
HydroxySphingomyelin C24:1
T
- TAG
Triacylglycerols
- TG(36:0)
Triglycerides(36:0)
- TG(56:11)
Triglycerides(56:11)
- Tyr
Tyrosine
V
- Val
Valine
Declarations
Conflict of interest
The authors have nothing to disclose.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Moloud Payab, Email: Moloudpayab@gmail.com.
Akram Tayanloo-Beik, Email: a.tayanloo@gmail.com.
Khadijeh Falahzadeh, Email: falahzadeh2020@yahoo.com.
Maryamossadat Mousavi, Email: mousavii.1990@gmail.com.
Saeede Salehi, Email: Sa.salehi123@gmail.com.
Shirin Djalalinia, Email: Shdjalalinia@gmail.com.
Mahbube Ebrahimpur, Email: mahbube10183@gmail.com.
Nafiseh Rezaei, Email: Nafis.rezaei@yahoo.com.
Mostafa Rezaei-Tavirani, Email: Tavirany@yahoo.com.
Bagher Larijani, Email: emrc@tums.ac.ir.
Babak Arjmand, Email: barjmand@sina.tums.ac.ir.
Kambiz Gilany, Email: k.gilany@ari.ir.
References
- 1.Abarca-Gómez L, Abdeen ZA, Hamid ZA, Abu-Rmeileh NM, Acosta-Cazares B, Acuin C, Adams RJ, Aekplakorn W, Afsana K, Aguilar-Salinas CA. Worldwide trends in body-mass index, underweight, overweight, and obesity from 1975 to 2016: a pooled analysis of 2416 population-based measurement studies in 128· 9 million children, adolescents, and adults. The Lancet. 2017;390:2627–2642. doi: 10.1016/S0140-6736(17)32129-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abu Bakar MH, Sarmidi MR, Cheng KK, Ali Khan A, Suan CL, Zaman Huri H, Yaakob H. Metabolomics - the complementary field in systems biology: a review on obesity and type 2 diabetes. Mol BioSyst. 2015;11:1742–1774. doi: 10.1039/c5mb00158g. [DOI] [PubMed] [Google Scholar]
- 3.Ahmad MS, Alsaleh M, Kimhofer T, Ahmad S, Jamal W, Wali SO, Nicholson JK, Damanhouri ZA, Holmes E. Metabolic zzphenotype of obesity in a Saudi population. J Proteome Res. 2017;16:635–644. doi: 10.1021/acs.jproteome.6b00710. [DOI] [PubMed] [Google Scholar]
- 4.Allam-Ndoul B, Guénard F, Garneau V, Cormier H, Barbier O, Pérusse L, Vohl M-C. Association between metabolite profiles, metabolic syndrome and obesity status. Nutrients. 2016;8:324. doi: 10.3390/nu8060324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Almanza-Aguilera E, Brunius C, Bernal-Lopez MR, Garcia-Aloy M, Madrid-Gambin F, Tinahones FJ, Gomez-Huelgas R, Landberg R, Andres-Lacueva C. Impact in plasma metabolome as effect of lifestyle intervention for weight-loss reveals metabolic benefits in metabolically healthy obese women. J Proteome Res. 2018;17:2600–2610. doi: 10.1021/acs.jproteome.8b00042. [DOI] [PubMed] [Google Scholar]
- 6.Andersen LW, Mackenhauer J, Roberts JC, Berg KM, Cocchi MN, Donnino MW. Etiology and therapeutic approach to elevated lactate levels. Mayo Clinic Proceedings. Elsevier; 2013. pp. 1127–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Badoud F, Lam KP, Dibattista A, Perreault M, Zulyniak MA, Cattrysse B, Stephenson S, Britz-Mckibbin P, Mutch DM. Serum and adipose tissue amino acid homeostasis in the metabolically healthy obese. J Proteome Res. 2014;13:3455–3466. doi: 10.1021/pr500416v. [DOI] [PubMed] [Google Scholar]
- 8.Badoud F, Perreault M, Zulyniak MA, Mutch DM. Molecular insights into the role of white adipose tissue in metabolically unhealthy normal weight and metabolically healthy obese individuals. FASEB J. 2015;29:748–758. doi: 10.1096/fj.14-263913. [DOI] [PubMed] [Google Scholar]
- 9.Baek SH, Kim M, Kim M, Kang M, Yoo HJ, Lee NH, Kim YH, Song M, Lee JH. Metabolites distinguishing visceral fat obesity and atherogenic traits in individuals with overweight. Obesity. 2017;25:323–331. doi: 10.1002/oby.21724. [DOI] [PubMed] [Google Scholar]
- 10.Bagheri M, Djazayery A, Qi L, Yekaninejad MS, Chamari M, Naderi M, Ebrahimi Z, Koletzko B, Uhl O, Farzadfar F. Effectiveness of vitamin D therapy in improving metabolomic biomarkers in obesity phenotypes: two randomized clinical trials. Int J Obes. 2018;42:1782–1796. doi: 10.1038/s41366-018-0107-0. [DOI] [PubMed] [Google Scholar]
- 11.Bagheri M, Djazayery A, Farzadfar F, Qi L, Yekaninejad MS, Aslibekyan S, Chamari M, Hassani H, Koletzko B, Uhl O. Plasma metabolomic profiling of amino acids and polar lipids in Iranian obese adults. Lipids Health Dis. 2019;18:1–9. doi: 10.1186/s12944-019-1037-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bagheri M, Djazayery A, Farzadfar F, Qi L, Yekaninejad MS, Aslibekyan S, Chamari M, Hassani H, Koletzko B, Uhl O. Plasma metabolomic profiling of amino acids and polar lipids in Iranian obese adults. Lipids Health Dis. 2019;18:94. doi: 10.1186/s12944-019-1037-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bakar MHA, Sarmidi MR, Cheng K-K, Khan AA, Suan CL, Huri HZ, Yaakob H. Metabolomics–the complementary field in systems biology: a review on obesity and type 2 diabetes. Mol BioSyst. 2015;11:1742–1774. doi: 10.1039/c5mb00158g. [DOI] [PubMed] [Google Scholar]
- 14.Brozinick J, Hawkins E, Bui HH, Kuo M, Tan B, Kievit P, Grove K. Plasma sphingolipids are biomarkers of metabolic syndrome in non-human primates maintained on a Western-style diet. Int J Obes. 2013;37:1064. doi: 10.1038/ijo.2012.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Butte NF, Liu Y, Zakeri IF, Mohney RP, Mehta N, Voruganti VS, Göring H, Cole SA, Comuzzie AG. Global metabolomic profiling targeting childhood obesity in the Hispanic population. Am J Clin Nutr. 2015;102:256–267. doi: 10.3945/ajcn.115.111872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cetin I, Parisi F, Berti C, Mando C, Desoye G. Placental fatty acid transport in maternal obesity. J Dev Orig Health Dis. 2012;3:409–414. doi: 10.1017/S2040174412000414. [DOI] [PubMed] [Google Scholar]
- 17.Chashmniam S, Madani NH, Ghoochani BFNM, Safari-Alighiarloo N, Khamseh ME. The metabolome profiling of obese and non-obese individuals: metabolically healthy obese and unhealthy non-obese paradox. Iranian J Basic Med Sci. 2020;23:186. doi: 10.22038/IJBMS.2019.37885.9004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen H-H, Tseng YJ, Wang S-Y, Tsai Y-S, Chang C-S, Kuo T-C, Yao W-J, Shieh C-C, Wu C-H, Kuo P-H. The metabolome profiling and pathway analysis in metabolic healthy and abnormal obesity. Int J Obes. 2015;39:1241. doi: 10.1038/ijo.2015.65. [DOI] [PubMed] [Google Scholar]
- 19.Cho K, Moon J, Kang JH, Jang H, Lee HJ, Park S, Yu KS, Cho JY. Combined untargeted and targeted metabolomic profiling reveals urinary biomarkers for discriminating obese from normal-weight adolescents. Pediatric obesity. 2017;12:93–101. doi: 10.1111/ijpo.12114. [DOI] [PubMed] [Google Scholar]
- 20.Collaborators, G. O Health effects of overweight and obesity in 195 countries over 25 years. N Engl J Med. 2017;377:13–27. doi: 10.1056/NEJMoa1614362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crawford SO, Hoogeveen RC, Brancati FL, Astor BC, Ballantyne CM, Schmidt MI, Young JH. Association of blood lactate with type 2 diabetes: the atherosclerosis risk in communities carotid MRI study. Int J Epidemiol. 2010;39:1647–1655. doi: 10.1093/ije/dyq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Desert R, Canlet C, Costet N, Cordier S, Bonvallot N. Impact of maternal obesity on the metabolic profiles of pregnant women and their offspring at birth. Metabolomics. 2015;11:1896–1907. [Google Scholar]
- 23.Dugas LR, Chorell E, Plange-Rhule J, Lambert EV, Cao G, Cooper RS, Layden BT, Scholten D, Olsson T, Luke A. Obesity-related metabolite profiles of black women spanning the epidemiologic transition. Metabolomics. 2016;12:45. doi: 10.1007/s11306-016-0960-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dunn WB, Lin W, Broadhurst D, Begley P, Brown M, Zelena E, Vaughan AA, Halsall A, Harding N, Knowles JD. Molecular phenotyping of a UK population: defining the human serum metabolome. Metabolomics. 2015;11:9–26. doi: 10.1007/s11306-014-0707-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elliott P, Posma JM, Chan Q, Garcia-Perez I, Wijeyesekera A, Bictash M, Ebbels TM, Ueshima H, Zhao L, van Horn L. Urinary metabolic signatures of human adiposity. Sci Transl Med. 2015;7:285ra62. doi: 10.1126/scitranslmed.aaa5680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fattuoni C, Mandò C, Palmas F, Anelli GM, Novielli C, Laudicina EP, Savasi VM, Barberini L, Dessì A, Pintus R. Preliminary metabolomics analysis of placenta in maternal obesity. Placenta. 2018;61:89–95. doi: 10.1016/j.placenta.2017.11.014. [DOI] [PubMed] [Google Scholar]
- 27.Feng R, Sun G, Zhang Y, Sun Q, Ju L, Sun C, Wang C. Short-term high-fat diet exacerbates insulin resistance and glycolipid metabolism disorders in young obese men with hyperlipidemia, as determined by metabolomics analysis using ultra-HPLC-quadrupole time-of-flight mass spectrometry. J Diabetes. 2019;11:148–160. doi: 10.1111/1753-0407.12828. [DOI] [PubMed] [Google Scholar]
- 28.Galili O, Versari D, Sattler KJ, Olson ML, Mannheim D, Mcconnell JP, Chade AR, Lerman LO, Lerman A. Early experimental obesity is associated with coronary endothelial dysfunction and oxidative stress. Am J Phys Heart Circ Phys. 2007;292:H904–H911. doi: 10.1152/ajpheart.00628.2006. [DOI] [PubMed] [Google Scholar]
- 29.Gault CR, Obeid LM, Hannun YA. An overview of sphingolipid metabolism: from synthesis to breakdown. Sphingolipids as Signaling and Regulatory Molecules. Springer; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gawlik A, Shmoish M, Hartmann MF, Malecka-Tendera E, Wudy SA, Hochberg ZE. Steroid metabolomic disease signature of nonsyndromic childhood obesity. J Clin Endocrinol Metab. 2016;101:4329–4337. doi: 10.1210/jc.2016-1754. [DOI] [PubMed] [Google Scholar]
- 31.Gibney MJ, Walsh M, Brennan L, Roche HM, German B, van Ommen B. Metabolomics in human nutrition: opportunities and challenges. Am J Clin Nutr. 2005;82:497–503. doi: 10.1093/ajcn.82.3.497. [DOI] [PubMed] [Google Scholar]
- 32.Hellmuth C, Demmelmair H, Schmitt I, Peissner W, Blüher M, Koletzko B. Association between plasma nonesterified fatty acids species and adipose tissue fatty acid composition. PLoS One. 2013;8:e74927. doi: 10.1371/journal.pone.0074927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hellmuth C, Lindsay KL, Uhl O, Buss C, Wadhwa PD, Koletzko B, Entringer S. Association of maternal prepregnancy BMI with metabolomic profile across gestation. Int J Obes. 2017;41:159. doi: 10.1038/ijo.2016.153. [DOI] [PubMed] [Google Scholar]
- 34.Hellmuth C, Uhl O, Standl M, Demmelmair H, Heinrich J, Koletzko B, Thiering E. Cord blood metabolome is highly associated with birth weight, but less predictive for later weight development. Obesity Facts. 2017;10:85–100. doi: 10.1159/000453001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hellmuth C, Lindsay KL, Uhl O, Buss C, Wadhwa PD, Koletzko B, Entringer S. Maternal Metabolomic profile and fetal programming of offspring adiposity: identification of potentially protective lipid metabolites. Mol Nutr Food Res. 2019;63:e1700889. doi: 10.1002/mnfr.201700889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ho JE, Larson MG, Ghorbani A, Cheng S, Chen M-H, Keyes M, Rhee EP, Clish CB, Vasan RS, Gerszten RE. Metabolomic profiles of body mass index in the Framingham heart study reveal distinct cardiometabolic phenotypes. PLoS One. 2016;11:e0148361. doi: 10.1371/journal.pone.0148361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Houten SM, Wanders RJ. A general introduction to the biochemistry of mitochondrial fatty acid β-oxidation. J Inherit Metab Dis. 2010;33:469–477. doi: 10.1007/s10545-010-9061-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hsu YH, Churchhouse C, Pers TH, Mercader JM, Metspalu A, Fischer K, Fortney K, Morgen EK, Gonzalez C, Gonzalez ME, Esko T, Hirschhorn JN. PAIRUP-MS: pathway analysis and imputation to relate unknowns in profiles from mass spectrometry-based metabolite data. PLoS Comput Biol. 2019;15:e1006734. doi: 10.1371/journal.pcbi.1006734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang C-F, Cheng M-L, Fan C-M, Hong C-Y, Shiao M-S. Nicotinuric acid: a potential marker of metabolic syndrome through a metabolomics-based approach. Diabetes Care. 2013;36:1729–1731. doi: 10.2337/dc12-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hulver MW, Berggren JR, Carper MJ, Miyazaki M, Ntambi JM, Hoffman EP, Thyfault JP, Stevens R, Dohm GL, Houmard JA. Elevated stearoyl-CoA desaturase-1 expression in skeletal muscle contributes to abnormal fatty acid partitioning in obese humans. Cell Metab. 2005;2:251–261. doi: 10.1016/j.cmet.2005.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iida m, Harada s, Kurihara A, Fukai K, Kuwabara K, Sugiyama D, Takeuchi A, Okamura T, Akiyama M, Nishiwaki Y. Profiling of plasma metabolites in postmenopausal women with metabolic syndrome. Menopause (New York, NY) 2016;23:749. doi: 10.1097/GME.0000000000000630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Isherwood CM, van der veen DR, Johnston JD, Skene DJ. Twenty-four-hour rhythmicity of circulating metabolites: effect of body mass and type 2 diabetes. FASEB J. 2017;31:5557–5567. doi: 10.1096/fj.201700323R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jonas A. Lecithin cholesterol acyltransferase. Biochimica et Biophysica Acta (BBA)-Molecular and Cell Biology of Lipids. 2000;1529:245–256. doi: 10.1016/s1388-1981(00)00153-0. [DOI] [PubMed] [Google Scholar]
- 44.Jourdan C, Petersen A-K, Gieger C, Döring A, Illig T, Wang-Sattler R, Meisinger C, Peters A, Adamski J, Prehn C. Body fat free mass is associated with the serum metabolite profile in a population-based study. PLoS One. 2012;7:e40009. doi: 10.1371/journal.pone.0040009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim JY, Park JY, Kim OY, Ham BM, Kim H-J, Kwon DY, Jang Y, Lee JH. Metabolic profiling of plasma in overweight/obese and lean men using ultra performance liquid chromatography and Q-TOF mass spectrometry (UPLC− Q-TOF MS) J Proteome Res. 2010;9:4368–4375. doi: 10.1021/pr100101p. [DOI] [PubMed] [Google Scholar]
- 46.Kim Y-J, Lee H-S, Kim YK, Park S, Kim J-M, Yun JH, Yu H-Y, Kim B-J. Association of metabolites with obesity and type 2 diabetes based on FTO genotype. PLoS One. 2016;11:e0156612. doi: 10.1371/journal.pone.0156612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim MJ, Kim JH, Kim MS, Yang HJ, Lee M, Kwon DY. Metabolomics associated with genome-wide association study related to the basal metabolic rate in overweight/obese Korean women. J Med Food. 2019;22:499–507. doi: 10.1089/jmf.2018.4310. [DOI] [PubMed] [Google Scholar]
- 48.Klautzer L, Becker J, Mattke S. The curse of wealth–middle eastern countries need to address the rapidly rising burden of diabetes. Int J Health Policy Manag. 2014;2:109. doi: 10.15171/ijhpm.2014.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klop B, Elte JWF, Cabezas MC. Dyslipidemia in obesity: mechanisms and potential targets. Nutrients. 2013;5:1218–1240. doi: 10.3390/nu5041218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kochhar S, Jacobs DM, Ramadan Z, Berruex F, Fuerholz A, Fay LB. Probing gender-specific metabolism differences in humans by nuclear magnetic resonance-based metabonomics. Anal Biochem. 2006;352:274–281. doi: 10.1016/j.ab.2006.02.033. [DOI] [PubMed] [Google Scholar]
- 51.Koletzko B, Beyer J, Brands B, Demmelmair H, Grote V, Haile G, Gruszfeld D, Rzehak P, Socha P, Weber M. Recent advances in growth research: nutritional, molecular and endocrine perspectives. Karger Publishers; 2013. Early influences of nutrition on postnatal growth. [DOI] [PubMed] [Google Scholar]
- 52.Koves TR, Ussher JR, Noland RC, Slentz D, Mosedale M, Ilkayeva O, Bain J, Stevens R, Dyck JR, Newgard CB. Mitochondrial overload and incomplete fatty acid oxidation contribute to skeletal muscle insulin resistance. Cell Metab. 2008;7:45–56. doi: 10.1016/j.cmet.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 53.Kupek E, Lobo AS, Leal DB, Bellisle F, de Assis MA. Dietary patterns associated with overweight and obesity among Brazilian schoolchildren: an approach based on the time-of-day of eating events. Br J Nutr. 2016;116:1954–1965. doi: 10.1017/S0007114516004128. [DOI] [PubMed] [Google Scholar]
- 54.Leal-Witt MJ, Ramon-Krauel M, Samino S, Llobet M, Cuadras D, Jimenez-Chillaron JC, Yanes O, Lerin C. Untargeted metabolomics identifies a plasma sphingolipid-related signature associated with lifestyle intervention in prepubertal children with obesity. Int J Obes. 2018;42:72–78. doi: 10.1038/ijo.2017.201. [DOI] [PubMed] [Google Scholar]
- 55.Lee SH, Kim SH, Lee W-Y, Chung BC, Park MJ, Choi MH. Metabolite profiling of sex developmental steroid conjugates reveals an association between decreased levels of steroid sulfates and adiposity in obese girls. J Steroid Biochem Mol Biol. 2016;162:100–109. doi: 10.1016/j.jsbmb.2016.04.020. [DOI] [PubMed] [Google Scholar]
- 56.Lei S, Huang F, Zhao A, Chen T, Chen W, Xie G, Zheng X, Zhang Y, Yu H, Zhang P. The ratio of dihomo-γ-linolenic acid to deoxycholic acid species is a potential biomarker for the metabolic abnormalities in obesity. FASEB J. 2017;31:3904–3912. doi: 10.1096/fj.201700055R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin Z, Gonçalves CMV, Dai L, Lu H-M, Huang J-H, Ji H, Wang D-S, Yi L-Z, Liang Y-Z. Exploring metabolic syndrome serum profiling based on gas chromatography mass spectrometry and random forest models. Anal Chim Acta. 2014;827:22–27. doi: 10.1016/j.aca.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 58.Lokhov PG, Balashova EE, Trifonova OP, Maslov DL, Ponomarenko EA, Archakov AI. Mass spectrometry-based metabolomics analysis of obese patients' blood plasma. Int J Mol Sci. 2020;21:568. doi: 10.3390/ijms21020568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mccormack SE, Shaham O, Mccarthy MA, Deik AA, Wang TJ, Gerszten RE, Clish CB, Mootha VK, Grinspoon SK, Fleischman A. Circulating branched-chain amino acid concentrations are associated with obesity and future insulin resistance in children and adolescents. Pediatric Obesity. 2013;8:52–61. doi: 10.1111/j.2047-6310.2012.00087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Menni C, Migaud M, Glastonbury CA, Beaumont M, Nikolaou A, Small KS, Brosnan MJ, Mohney RP, Spector TD, Valdes AM. Metabolomic profiling to dissect the role of visceral fat in cardiometabolic health. Obesity. 2016;24:1380–1388. doi: 10.1002/oby.21488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mihalik SJ, Goodpaster BH, Kelley DE, Chace DH, Vockley J, Toledo FG, Delany JP. Increased levels of plasma acylcarnitines in obesity and type 2 diabetes and identification of a marker of glucolipotoxicity. Obesity. 2010;18:1695–1700. doi: 10.1038/oby.2009.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mihalik SJ, Michaliszyn SF, De Las Heras J, Bacha F, Lee S, Chace DH, Dejesus VR, Vockley J, Arslanian SA. Metabolomic profiling of fatty acid and amino acid metabolism in youth with obesity and type 2 diabetes: evidence for enhanced mitochondrial oxidation. Diabetes Care. 2012:DC_111577. [DOI] [PMC free article] [PubMed]
- 63.Murphy RA, Moore SC, Playdon M, Meirelles O, Newman AB, Milijkovic I, Kritchevsky SB, Schwartz A, Goodpaster BH, Sampson J. Metabolites associated with lean mass and adiposity in older black men. Journals of Gerontology Series A: Biomedical Sciences and Medical Sciences. 2017;72:1352–1359. doi: 10.1093/gerona/glw245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Newbern D, Balikcioglu PG, Balikcioglu M, Bain J, Muehlbauer M, Stevens R, Ilkayeva O, Dolinsky D, Armstrong S, Irizarry K. Sex differences in biomarkers associated with insulin resistance in obese adolescents: metabolomic profiling and principal components analysis. J Clin Endocrinol Metab. 2014;99:4730. doi: 10.1210/jc.2014-2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Okekunle AP, Li Y, Liu L, Du S, Wu X, Chen Y, Li Y, Qi J, Sun C, Feng R. Abnormal circulating amino acid profiles in multiple metabolic disorders. Diabetes Res Clin Pract. 2017;132:45–58. doi: 10.1016/j.diabres.2017.07.023. [DOI] [PubMed] [Google Scholar]
- 66.Organization, W. H . Global status report on noncommunicable diseases 2014. World Health Organization; 2014. [Google Scholar]
- 67.Palau-Rodriguez M, Tulipani S, Marco-Ramell A, Minarro A, Jauregui O, Gonzalez-Dominguez R, Sanchez-Pla A, Ramos-Molina B, Tinahones FJ, Andres-Lacueva C. Characterization of metabolomic profile associated with metabolic improvement after bariatric surgery in subjects with morbid obesity. J Proteome Res. 2018;17:2704–2714. doi: 10.1021/acs.jproteome.8b00144. [DOI] [PubMed] [Google Scholar]
- 68.Palmnas MSA, Kopciuk KA, Shaykhutdinov RA, Robson PJ, Mignault D, Rabasa-Lhoret R, Vogel HJ, Csizmadi I. Serum metabolomics of activity energy expenditure and its relation to metabolic syndrome and obesity. Sci Rep. 2018;8:3308. doi: 10.1038/s41598-018-21585-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Paris D, Maniscalco M, Melck D, D’Amato M, Sorrentino N, Zedda A, Sofia M, Motta A. Inflammatory metabolites in exhaled breath condensate characterize the obese respiratory phenotype. Metabolomics. 2015;11:1934–1939. [Google Scholar]
- 70.Park S, Sadanala KC, KIM E-K. A metabolomic approach to understanding the metabolic link between obesity and diabetes. Mol Cells. 2015;38:587. doi: 10.14348/molcells.2015.0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Payab M, Hasani-Ranjbar S, Larijani B. whether all obese subjects both in metabolic groups and non-metabolic groups should be treated or not. springer; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Payab M, Kelishadi R, Qorbani M, Motlagh ME, Ranjbar SH, Ardalan G, Zahedi H, Chinian M, Asayesh H, Larijani B. Association of junk food consumption with high blood pressure and obesity in Iranian children and adolescents: the Caspian-IV study. Jornal de Pediatria (Versão em Português) 2015;91:196–205. doi: 10.1016/j.jped.2014.07.006. [DOI] [PubMed] [Google Scholar]
- 73.Payab M, Kelishadi R, Ranjbar SH, Motlagh ME, Ardalan G, Zahedi H, Sanaei M, Shafiee G, Asayesh H, Larijani B. Grains and potato consumption in association with anthropomet¬ ric measures and blood pressure in Iranian Chil¬ dren and adolescents: the CASPIAN-IV study. Iran J Public Health. 2015;44:25–34. [Google Scholar]
- 74.Perng W, Gillman MW, Fleisch AF, Michalek RD, Watkins SM, Isganaitis E, Patti ME, Oken E. Metabolomic profiles and childhood obesity. Obesity. 2014;22:2570–2578. doi: 10.1002/oby.20901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Phung DT, Wang Z, Rutherford S, Huang C, Chu C. Body mass index and risk of pneumonia: a systematic review and meta-analysis. Obes Rev. 2013;14:839–857. doi: 10.1111/obr.12055. [DOI] [PubMed] [Google Scholar]
- 76.Pietiläinen KH, Sysi-Aho M, Rissanen A, Seppänen-Laakso T, Yki-Järvinen H, Kaprio J, Orešič M. Acquired obesity is associated with changes in the serum lipidomic profile independent of genetic effects–a monozygotic twin study. PLoS One. 2007;2:e218. doi: 10.1371/journal.pone.0000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Putri SP, Yamamoto S, Tsugawa H, Fukusaki E. Current metabolomics: technological advances. J Biosci Bioeng. 2013;116:9–16. doi: 10.1016/j.jbiosc.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 78.Rangel-Huerta OD, Gil A. Nutrimetabolomics: an update on analytical approaches to investigate the role of plant-based foods and their bioactive compounds in non-communicable chronic diseases. Int J Mol Sci. 2016;17:2072. doi: 10.3390/ijms17122072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rauschert S, Uhl O, Koletzko B, Kirchberg F, Mori TA, Huang R-C, Beilin LJ, Hellmuth C, Oddy WH. Lipidomics reveals associations of phospholipids with obesity and insulin resistance in young adults. J Clin Endocrinol Metab. 2016;101:871–879. doi: 10.1210/jc.2015-3525. [DOI] [PubMed] [Google Scholar]
- 80.Rauschert S, Mori TA, Beilin LJ, Jacoby p, Uhl o, Koletzko B, Oddy WH, Hellmuth C. Early life factors, obesity risk, and the metabolome of young adults. Obesity. 2017;25:1549–1555. doi: 10.1002/oby.21915. [DOI] [PubMed] [Google Scholar]
- 81.Romo-Hualde A, Huerta AE, Gonzalez-Navarro CJ, Ramos-Lopez O, Moreno-Aliaga MJ, Martinez JA. Untargeted metabolomic on urine samples after alpha-lipoic acid and/or eicosapentaenoic acid supplementation in healthy overweight/obese women. Lipids Health Dis. 2018;17:103. doi: 10.1186/s12944-018-0750-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rousset X, Vaisman B, Amar M, Sethi AA, Remaley AT. Lecithin: cholesterol acyltransferase: from biochemistry to role in cardiovascular disease. Curr Opin Endocrinol Diabetes Obes. 2009;16:163. doi: 10.1097/med.0b013e328329233b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sallese A, Zhu J. Mass spectrometry based metabolomics: A novel analytical technique for detecting metabolic syndrome? Bioanalysis. 2017;9:1623–1626. doi: 10.4155/bio-2017-0165. [DOI] [PubMed] [Google Scholar]
- 84.Sandler V, Reisetter AC, Bain JR, Muehlbauer MJ, Nodzenski M, Stevens RD, Ilkayeva O, Lowe LP, Metzger BE, Newgard CB. Associations of maternal BMI and insulin resistance with the maternal metabolome and newborn outcomes. Diabetologia. 2017;60:518–530. doi: 10.1007/s00125-016-4182-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schlecht I, Gronwald W, Behrens G, Baumeister SE, Hertel J, Hochrein J, Zacharias HU, Fischer B, Oefner PJ, Leitzmann MF. Visceral adipose tissue but not subcutaneous adipose tissue is associated with urine and serum metabolites. PLoS ONE. 2017;12:e0175133. doi: 10.1371/journal.pone.0175133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schoeman JC, Hou J, Harms AC, Vreeken RJ, Berger R, Hankemeier T, Boonstra A. Metabolic characterization of the natural progression of chronic hepatitis B. Genome Med. 2016;8:64. doi: 10.1186/s13073-016-0318-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Seridi L, Leo GC, Dohm GL, Pories WJ, Lenhard J. Time course metabolome of roux-en-Y gastric bypass confirms correlation between leptin, body weight and the microbiome. PLoS One. 2018;13:e0198156. doi: 10.1371/journal.pone.0198156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shokry E, Marchioro L, Uhl O, Bermudez MG, Garcia-Santos JA, Segura MT, Campoy C, Koletzko B. Impact of maternal BMI and gestational diabetes mellitus on maternal and cord blood metabolome: results from the PREOBE cohort study. Acta Diabetol. 2019;56:421–430. doi: 10.1007/s00592-019-01291-z. [DOI] [PubMed] [Google Scholar]
- 89.Subbaiah PV, Jiang X-C, Belikova NA, Aizezi B, Huang ZH, Reardon CA. Regulation of plasma cholesterol esterification by sphingomyelin: effect of physiological variations of plasma sphingomyelin on lecithin-cholesterol acyltransferase activity. Biochimica et Biophysica Acta (BBA)-Molecular and Cell Biology of Lipids. 2012;1821:908–913. doi: 10.1016/j.bbalip.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sun L, Hu C, Yang R, Lv Y, Yuan H, Liang Q, He B, Pang G, Jiang M, Dong J, Yang Z. Association of circulating branched-chain amino acids with cardiometabolic traits differs between adults and the oldest-old. Oncotarget. 2017;8:88882–88893. doi: 10.18632/oncotarget.21489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Troisi J, Belmonte F, Bisogno A, Pierri L, Colucci A, Scala G, Cavallo P, Mandato C, Di Nuzzi A, Di Michele L, Delli Bovi AP, Guercio Nuzio S, Vajro P. Metabolomic Salivary Signature of Pediatric Obesity Related Liver Disease and Metabolic Syndrome. Nutrients. 2019:11. [DOI] [PMC free article] [PubMed]
- 92.Tulipani S, Palau-Rodriguez M, Alonso AM, Cardona F, Marco-Ramell A, Zonja B, De Alda ML, Muñoz-Garach A, Sanchez-Pla A, Tinahones FJ. Biomarkers of morbid obesity and prediabetes by metabolomic profiling of human discordant phenotypes. Clin Chim Acta. 2016;463:53–61. doi: 10.1016/j.cca.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 93.Valcárcel B, Ebbels TM, Kangas AJ, Soininen P, Elliot P, Ala-Korpela M, Järvelin M-R, De Iorio M. Genome metabolome integrated network analysis to uncover connections between genetic variants and complex traits: an application to obesity. J R Soc Interface. 2014;11:20130908. doi: 10.1098/rsif.2013.0908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vijay A, Valdes AM. The Metabolomic signatures of weight change. Metabolites. 2019;9:67. doi: 10.3390/metabo9040067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vitkin E, Ben-Dor A, Shmoish M, Hartmann MF, Yakhini Z, Wudy SA, Hochberg ZE. Peer group normalization and urine to blood context in steroid metabolomics: the case of CAH and obesity. Steroids. 2014;88:83–89. doi: 10.1016/j.steroids.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 96.Wahl S, Yu Z, Kleber M, Singmann P, Holzapfel C, He Y, Mittelstrass K, Polonikov A, Prehn C, Römisch-Margl W. Childhood obesity is associated with changes in the serum metabolite profile. Obesity Facts. 2012;5:660–670. doi: 10.1159/000343204. [DOI] [PubMed] [Google Scholar]
- 97.Wahl S, Drong A, Lehne B, Loh M, Scott WR, Kunze S, Tsai PC, Ried JS, Zhang W, Yang Y, Tan S, Fiorito G, Franke L, Guarrera S, Kasela S, Kriebel J, Richmond RC, Adamo M, Afzal U, Ala-Korpela M, Albetti B, Ammerpohl O, Apperley JF, Beekman M, Bertazzi PA, Black SL, Blancher C, Bonder MJ, Brosch M, Carstensen-Kirberg M, De Craen AJ, De Lusignan S, Dehghan A, Elkalaawy M, Fischer K, Franco OH, Gaunt TR, Hampe J, Hashemi M, Isaacs A, Jenkinson A, Jha S, Kato N, Krogh V, Laffan M, Meisinger C, Meitinger T, Mok ZY, Motta V, Ng HK, Nikolakopoulou Z, Nteliopoulos G, Panico S, Pervjakova N, Prokisch H, Rathmann W, Roden M, Rota F, Rozario MA, Sandling JK, Schafmayer C, Schramm K, Siebert R, Slagboom PE, Soininen P, Stolk L, Strauch K, Tai ES, Tarantini L, Thorand B, Tigchelaar EF, Tumino R, Uitterlinden AG, Van Duijn C, Van Meurs JB, Vineis P, Wickremasinghe AR, Wijmenga C, Yang TP, Yuan W, Zhernakova A, Batterham RL, Smith GD, Deloukas P, Heijmans BT, Herder C, Hofman A, Lindgren CM, Milani L, Van Der Harst P, Peters A, Illig T, Relton CL, Waldenberger M, Jarvelin MR, Bollati V, Soong R, Spector TD, Scott J, Mccarthy MI, et al. Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity. Nature. 2017;541:81–86. doi: 10.1038/nature20784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang C, Feng R, Sun D, Li Y, Bi X, Sun C. Metabolic profiling of urine in young obese men using ultra performance liquid chromatography and Q-TOF mass spectrometry (UPLC/Q-TOF MS) J Chromatogr B. 2011;879:2871–2876. doi: 10.1016/j.jchromb.2011.08.014. [DOI] [PubMed] [Google Scholar]
- 99.Wang Y, Liu D, Li Y, Guo L, Cui Y, Zhang X, Li E. Metabolomic analysis of serum from obese adults with hyperlipemia by UHPLC-Q-TOF MS/MS. Biomed Chromatogr. 2016;30:48–54. doi: 10.1002/bmc.3491. [DOI] [PubMed] [Google Scholar]
- 100.Wang SM, Yang RY, Wang M, Ji FS, Li HX, Tang YM, Chen WX, Dong J. Identification of serum metabolites associated with obesity and traditional risk factors for metabolic disease in Chinese adults. Nutr Metab Cardiovasc Dis. 2018;28:112–118. doi: 10.1016/j.numecd.2017.09.009. [DOI] [PubMed] [Google Scholar]
- 101.Wijayatunga NN, Sams VG, Dawson JA, Mancini ML, Mancini GJ, Moustaid-Moussa N. Roux-en-Y gastric bypass surgery alters serum metabolites and fatty acids in patients with morbid obesity. Diabetes Metab Res Rev. 2018;34:e3045. doi: 10.1002/dmrr.3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Würtz P, Wang Q, Kangas AJ, Richmond RC, Skarp J, Tiainen M, Tynkkynen T, soininen P, Havulinna AS, Kaakinen M. Metabolic signatures of adiposity in young adults: Mendelian randomization analysis and effects of weight change. PLoS Med. 2014;11:e1001765. doi: 10.1371/journal.pmed.1001765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Xia B, Zhu Q, Zhao Y, Ge W, Zhao Y, Song Q, Zhou Y, Shi H, Zhang Y. Phthalate exposure and childhood overweight and obesity: urinary metabolomic evidence. Environ Int. 2018;121:159–168. doi: 10.1016/j.envint.2018.09.001. [DOI] [PubMed] [Google Scholar]
- 104.Xie G, Ma X, Zhao A, Wang C, Zhang Y, Nieman D, Nicholson JK, Jia W, Bao Y, Jia W. The metabolite profiles of the obese population are gender-dependent. J Proteome Res. 2014;13:4062–4073. doi: 10.1021/pr500434s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yin X, Subramanian S, Willinger CM, Chen G, Juhasz P, Courchesne P, Chen BH, Li X, Hwang S-J, Fox CS. Metabolite signatures of metabolic risk factors and their longitudinal changes. J Clin Endocrinol Metab. 2016;101:1779–1789. doi: 10.1210/jc.2015-2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yu HT, Fu XY, Xu B, Zuo LL, Ma HB, Wang SR. Untargeted metabolomics approach (UPLC-Q-TOF-MS) explores the biomarkers of serum and urine in overweight/obese young men. Asia Pac J Clin Nutr. 2018;27:1067–1076. doi: 10.6133/apjcn.052018.07. [DOI] [PubMed] [Google Scholar]
- 107.Zhang A, Sun H, Wang X. Emerging role and recent applications of metabolomics biomarkers in obesity disease research. RSC Adv. 2017;7:14966–14973. [Google Scholar]
- 108.Zhao Q, Zhu Y, Best LG, Umans JG, Uppal K, Tran VT, Jones DP, Lee ET, Howard BV, Zhao J. Metabolic profiles of obesity in American Indians: the strong heart family study. PLoS ONE. 2016;11:e0159548. doi: 10.1371/journal.pone.0159548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhong F, Xu M, Bruno RS, Ballard KD, Zhu J. Targeted high performance liquid chromatography tandem mass spectrometry-based metabolomics differentiates metabolic syndrome from obesity. Exp Biol Med. 2017;242:773–780. doi: 10.1177/1535370217694098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(RAR 529 kb)