ABSTRACT
The cellular and genetic mechanisms that coordinate formation of facial sensory structures with surrounding skeletal and soft tissue elements remain poorly understood. Alx1, a homeobox transcription factor, is a key regulator of midfacial morphogenesis. ALX1 mutations in humans are linked to severe congenital anomalies of the facial skeleton (frontonasal dysplasia, FND) with malformation or absence of eyes and orbital contents (micro- and anophthalmia). Zebrafish with loss-of-function alx1 mutations develop with craniofacial and ocular defects of variable penetrance, likely due to compensatory upregulation in expression of a paralogous gene, alx3. Here we show that zebrafish alx1;alx3 mutants develop with highly penetrant cranial and ocular defects that resemble human ALX1-linked FND. alx1 and alx3 are expressed in anterior cranial neural crest (aCNC), which gives rise to the anterior neurocranium (ANC), anterior segment structures of the eye and vascular pericytes. Consistent with a functional requirement for alx genes in aCNC, alx1; alx3 mutants develop with nearly absent ANC and grossly aberrant hyaloid vasculature and ocular anterior segment, but normal retina. In vivo lineage labeling identified a requirement for alx1 and alx3 during aCNC migration, and transcriptomic analysis suggested oxidative stress response as a key target mechanism of this function. Oxidative stress is a hallmark of fetal alcohol toxicity, and we found increased penetrance of facial and ocular malformations in alx1 mutants exposed to ethanol, consistent with a protective role for alx1 against ethanol toxicity. Collectively, these data demonstrate a conserved role for zebrafish alx genes in controlling ocular and facial development, and a novel role in protecting these key midfacial structures from ethanol toxicity during embryogenesis. These data also reveal novel roles for alx genes in ocular anterior segment formation and vascular development and suggest that retinal deficits in alx mutants may be secondary to aberrant ocular vascularization and anterior segment defects. This study establishes robust zebrafish models for interrogating conserved genetic mechanisms that coordinate facial and ocular development, and for exploring gene–environment interactions relevant to fetal alcohol syndrome.
KEY WORDS: Neural crest, ALX, Frontonasal dysplasia, Ocular anterior segment, Fetal alcohol syndrome
Summary: We show that zebrafish alx1 and alx3 function redundantly to regulate ocular and facial development, and identify a novel protective function of alx1 against ethanol toxicity.
INTRODUCTION
The vertebrate face comprises ocular, vascular and skeletal cell lineages that are closely apposed during development. The anterior cranial neural crest (aCNC) originates from the anterior–dorsal neural tube (future forebrain and midbrain), migrates cephalad to the eyes and turns caudally toward the nascent mouth opening to give rise to facial chondrocytes (Kague et al., 2012; Kish et al., 2011; Kuratani, 2018; Serbedzija et al., 1992; Wada et al., 2005), ocular anterior segments, e.g. the cornea and iris (Cavodeassi et al., 2019; Cvekl and Tamm, 2004; Ma and Lwigale, 2019; Williams and Bohnsack, 2015), and vascular pericytes of ocular blood vessels (Gage et al., 2005; Trost et al., 2013). Despite their importance, the gene networks that regulate these aCNC-derived lineages are poorly understood.
The Aristaless-like (alx) gene family consists of ALX1, ALX3 and ALX4 in human, mouse and zebrafish; ALX4 is represented by duplicated alx4a and alx4b in zebrafish (Dee et al., 2013). In humans, ALX1 mutations are linked to severe frontonasal dysplasia (FND) and extreme microphthalmia, and ALX3 and ALX4 genes are associated with a phenotypic spectrum of hypertelorism and nasal-tip duplications (Pini et al., 2020; Uz et al., 2010). These functions of alx genes are conserved in other vertebrates, but the pathogenic developmental mechanisms leading to the observed craniofacial malformations remain largely unknown (Beverdam et al., 2001; Dee et al., 2013; Forsthoefel, 1963; Lakhwani et al., 2010; Lyons et al., 2016; Pini et al., 2020; Qu et al., 1999; Zhao et al., 1996). In mice, disruption of alx1 results in anencephaly, precluding analysis of its role in facial and ocular morphogenesis, while compound mutants of alx3 and alx4 present with FND-like defects (Beverdam et al., 2001; Lakhwani et al., 2010).
Mouse alx family members, alx1, alx3 and alx4, are co-expressed in aCNC that gives rise to the anterior neurocranium (ANC), the primary palate (Dee et al., 2013; McGonnell et al., 2011; Mitchell et al., 2021; Qu et al., 1999; ten Berge et al., 1998), anterior segment and periocular neural crest (Ma and Lwigale, 2019; Sedykh et al., 2017). Recent reports demonstrate that zebrafish alx1 is required for regulating neural crest differentiation and migration in vitro and in vivo (Pini et al., 2020) and that zebrafish alx3 controls ANC chondrocyte differentiation in zebrafish (Mitchell et al., 2021). However, these studies do not address the role for alx function in the developing eye, nor do they examine the potential for compensatory interactions between alx1 and alx3.
While hereditary FND is rare, similar facial and ocular malformations are commonly associated with fetal alcohol exposure in humans (Farlie et al., 2016; Lovely et al., 2017). Genetic susceptibility to ethanol toxicity is actively studied in model organisms (Boschen et al., 2021; de la Morena-Barrio et al., 2018; Eberhart and Parnell, 2016; Kaminen-Ahola, 2020; Lovely et al., 2017), but much remains to be learned about the mechanisms of gene–environment interactions, particularly at the level of transcription.
Here we report that ALX1 homozygous loss-of-function mutation is associated with severe ocular malformation in humans and show that alx1 mutant fish also develop with ocular impairments. Targeted mutagenesis of a paralogous gene, alx3, was carried out to generate alx1;alx3 double-mutant zebrafish and to demonstrate that variable penetrance of alx1 mutant defects in zebrafish is due to functional redundancy with alx3. Our data demonstrate genetic requirements for alx1 and alx3 in craniofacial cartilage development, at least in part through regulating migratory aCNC that gives rise to the median element of the ANC. Importantly, we report evidence of unexpected roles for alx1 and alx3 in ocular vasculature and identify a role for alx1 and alx3 in regulating oxidative stress response and ribosome biogenesis. Consistent with these findings, zebrafish alx1 protects ocular and facial embryonic lineages against the effects of ethanol toxicity, which acts through ethanol-induced oxidative stress in neural crest cells. Collectively, these data establish the alx1 and alx1;alx3 zebrafish models as unique new tools for dissecting both the genetic controls and gene-environment interactions during craniofacial and ocular development.
RESULTS
Human ALX1 and zebrafish alx1 are required for ocular development
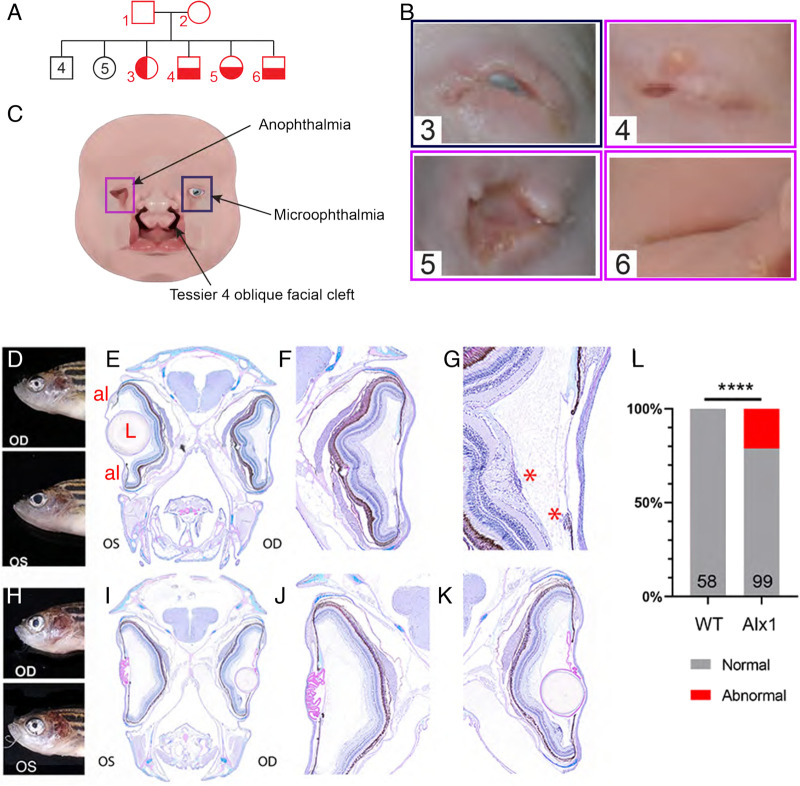
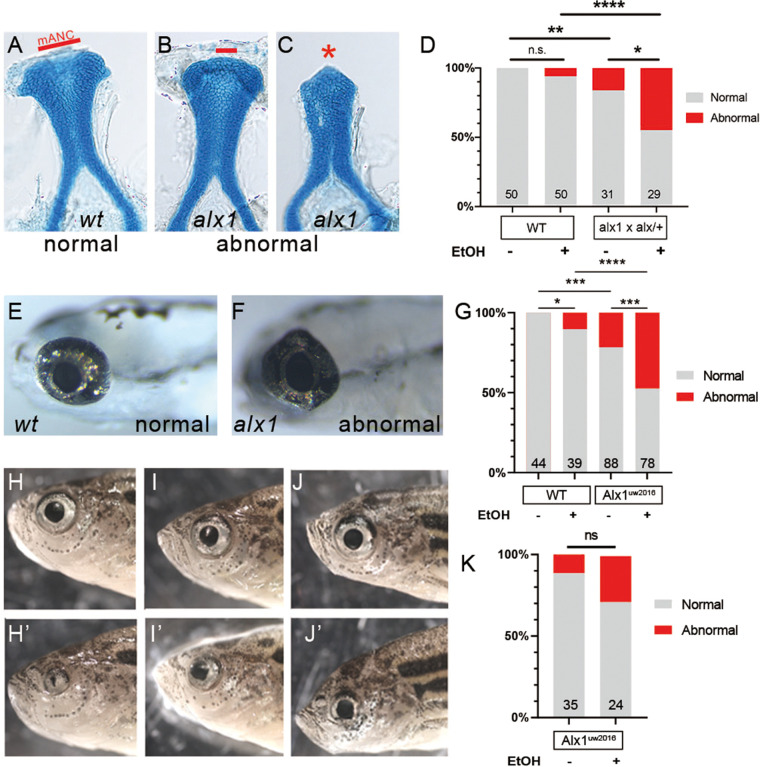
We recently reported a pedigree with consanguineous parents and 13 children, four of whom were born with FND linked to a missense gene variant, l165f, in the homeodomain of alx1 (Pini et al., 2020) (Fig. 1A). All four ALX1-homozygous patients presented with oblique orofacial clefts together with severe bilateral ocular deficits (Fig. 1B,C). These findings are consistent with a previous study that described patients with deletions that encompassed ALX1 and other loci, and a patient with splice-site mutation at the ALX1 locus (Uz et al., 2010). While the mechanism of ALX1 function in the facial cartilage is coming to light through active investigation, the role for ALX1 during ocular development remains unknown. We sought to address this knowledge gap by establishing zebrafish models of ALX1-linked FND. Using CRISPR/Cas9-mediated targeted mutagenesis, we generated loss-of-function alleles in zebrafish alx1 and showed that the majority of alx1 homozygotes were viable as adults, with a small proportion exhibiting craniofacial malformations that manifest during larval stages (Pini et al., 2020). On gross anatomical examination, we observed a range of ocular impairments in alx1uw2016 adults. Histological analysis of four representative affected adults revealed aberrant pupils, missing lenses (aphakia) and iridial dysplasia (Fig. 1D-K), but largely normal retinae. These ocular anomalies occurred in at least one eye in a significant proportion of alx1uw2016 adults (21 of 99 total, P<0.0001, Fig. 1L). These findings suggest a post-embryonic requirement for zebrafish alx1 function, which may be restricted to the anterior segment lineages of the eye.
Fig. 1.
Ocular malformations are associated with ALX1/alx1 loss-of-function in humans and zebrafish. (A) Consanguineous ALX1L165F/+ parents produced 13 children, four of whom were homozygous for ALX1L165F and had complex frontonasal dysplasia (FND3) (subject numbers noted in red). Unaffected individuals did not have eye or facial phenotypes suggestive of FND. (B) Diagram summarizing the anophthalmia, coloboma and bilateral Tessier oblique facial clefts (ObFC) characteristic of FND3. (C) Human subjects displayed bilateral ObFC (not shown) and a range of ocular malformations. The eldest sibling (subject 3) presented with right coloboma and left microophthalmia. The next child (subject 4) presented with bilateral anophthalmia with fused eyelids and shallow orbits. Subject 5 presented with bilateral anophthalmia with open shallow orbits. The upper and lower eyelids were absent, exposing the orbital mucosa. The nasal alae were also malformed with nodular skin tags. Subject 6 had bilateral anophthalmia, fused eyelids, and shallow orbit, similar to subject 4. (D-L) alx1uw2016 adult zebrafish exhibit milder ocular defects, primarily in the anterior ocular segment. (D-G) A representative alx1uw2016 adult with a unilateral ocular malformation (D). (E) Transverse section through the head of the fish in E shows a normal left eye and aphakia (absent lens) on the right. (F) The right eye lacks the lens and the ventral annular ligament but is largely normal otherwise. (G) The right eye contains mononuclear inflammatory cells near the ventral iris and the optic nerve head (asterisks). (H-K) A representative alx1uw2016 zebrafish with bilateral ocular defects (H). (J) In transverse section through the head, the left eye is missing the lens (aphakia) and contains a thick, multifocally disrupted lens capsule, which is embedded in the anterior chamber. There is no annular ligament on the dorsal iridocorneal angle. (K) The right eye contains a small spherical lens nucleus inside a wrinkled lens capsule. The ventral iridocorneal angle is devoid of the annular ligament. (L) Ocular defects were observed in 21.2% of alx1uw2016 homozygotes examined, and none of the wildtype fish of a similar age (****P<0.0001, Fisher's exact test). OD: oculus dexter, right eye; OS: oculus sinister, left eye; al: annular ligament; L: lens; *, mononuclear inflammatory cells.
Zebrafish alx1 and alx3 function redundantly during craniofacial lineage formation
alx1 and its homologs, alx3 and alx4, are co-expressed in aCNC, which gives rise to the median element of the ANC (Dee et al., 2013; McGonnell et al., 2011; Mitchell et al., 2021; Qu et al., 1999; ten Berge et al., 1998), as well as to the ocular anterior segment and periocular neural crest (Ma and Lwigale, 2019; Sedykh et al., 2017). All four single-locus mutants have been analyzed but only alx1 mutants (Pini et al., 2020) and alx3 mutants (Mitchell et al., 2021) exhibited embryonic defects. These defects were far milder than those reported in antisense morpholino-mediated knockdown assays (Dee et al., 2013) and did not recapitulate the severe craniofacial and ocular phenotypes in ALX1-linked (Pini et al., 2020; Uz et al., 2010) or ALX3-linked (Twigg et al., 2009) FND patients. These findings point to redundant or compensatory functions of alx genes in the embryonic zebrafish.
A recent study used single cell RNA-seq to show that all four zebrafish alx genes are expressed in frontonasal neural crest at 24 hours post fertilization (hpf) (Mitchell et al., 2021). We have also shown that alx3 transcription is increased in alx1 mutants (Pini et al., 2020), likely through the process of transcriptional adaptation (El-Brolosy et al., 2019). We hypothesized that the low penetrance of craniofacial malformations in alx1 mutants is due to functional compensation by alx3 and used CRISPR/Cas9 mutagenesis to mutagenize alx3 in the alx1uw2016 background (see Materials and Methods). Three frame-shift mutant alleles were recovered; since homozygotes for each allele developed with similar phenotypes, we chose one of the alleles, a 13-nucleotide insertion allele named alx3uw2113, for in-depth analysis. alx3uw2113 is predicted to encode a non-functional protein due to premature translation termination that results from a frame-shift mutation upstream of the conserved homeodomain and transactivation domains of alx3 (Fig. S1).
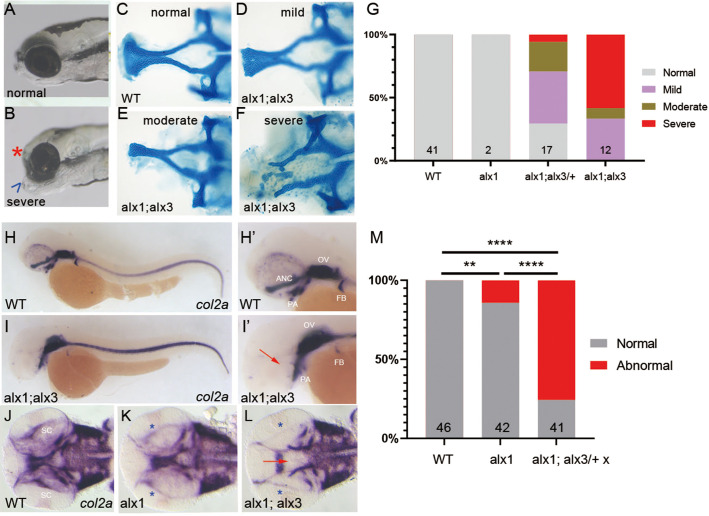
alx1uw2016;alx3uw2113/+ and alx1uw2016;alx3uw2113 (abbreviated alx1;alx3/+ and alx1;alx3) zebrafish were viable as adults and fertile, with alx1;alx3 adults exhibiting a range of facial anomalies (data not shown). alx1;alx3 larvae developed with a distinctive facial phenotype consistent with hypoplastic ANC at 5 days post-fertilization (dpf) (Fig. 2A,B). Alcian Blue staining, which labels neural crest-derived chondrocytes, confirmed ANC hypoplasia with various degrees of severity in the mutant larvae (Fig. 2C-F). Notably, alx1;alx3/+ embryos also developed with anomalous ANC primordia, albeit milder and at a lower penetrance (Fig. 2G).
Fig. 2.
alx1 and alx3 function redundantly to control anterior neurocranium morphogenesis in zebrafish. Larvae derived from a cross between alx1;alx3/+ parents were fixed at 5 dpf. (A,B) Representative siblings with normal facial structures (A) and with aberrantly short neurocranium (asterisk) and abnormally protruding jaw cartilage (arrowhead; B). (C-F) Alcian Blue staining revealed EP anomalies that range from mild (reduced median ethmoid plate width in D), moderate (absent median and reduced lateral ethmoid plate in E), to severe (absent ethmoid plate in F). (G) Penetrance and severity of cartilage defects is increased in alx1;alx3 compared to alx1;alx3/+ embryos (two trials). (H-M) Embryos derived from wildtype, alx1, or alx1;alx3/+ parental crosses were fixed at 2 dpf and stained for col2a1a expression using whole-mount in situ hybridization. (H,H′) Normal col2a1a expression in a wildtype embryo. (I,I′) A truncated EP in an alx1;alx3 embryo (arrow). (J) Normal col2a1a expression in a wildtype embryo. (K) Normal ethmoid plate and reduced presumptive scleral staining (asterisk) in an alx1 mutant. (L) Hypoplastic ethmoid plate (arrow) and absent presumptive scleral staining (asterisk) in an alx1;alx3 mutant. ANC, anterior neurocranium; OV, optic vesicle; PA, pharyngeal arches; FB, fin bud; SC, sclera. M: 75% of the embryos derived from alx1;alx3/+ have aberrant ANC and reduced scleral precursors (**P=0.0097, ****P<0.0001, Fisher's exact test; two trials for wildtype and alx1;alx3/+, and one trial for alx1). Embryos in A,B, and H-I′ are shown in lateral views, anterior to the left. Embryos in J-L are shown in ventral views, anterior to the left.
We next sought to define the earliest time that craniofacial cartilage defects manifest in alx1;alx3 embryos. To do this, we examined expression of col2a1a, an early chondrocyte marker (Yan et al., 1995) in wildtype, alx1uw2016 (abbreviated alx1), and alx1;alx3 embryos at 2 dpf. The majority of alx1 embryos (88%) exhibited normal expression of col2a1a (Fig. 2H,M), consistent with the low penetrance of deficits by 5 dpf. In contrast, only 24% of embryos derived from an alx1;alx3/+ incross developed with normal ANC, while 76% had truncated or missing ANC at 5 dpf (Fig. 2I,I′,L,M). This phenotypic ratio indicates that the ANC primordium is aberrant in both alx1;alx3 and alx1;alx3/+ embryos by 2 dpf. col2a1a expression was also detected in the presumptive sclera, the cartilaginous outer shell of the eye globe contiguous with the cornea, which is derived from neural crest (Gestri et al., 2012). col2a1a expression in putative scleral precursors was reduced in alx1 and alx1;alx3 embryos (Fig. 2K,L), suggesting an early requirement for alx1 and alx3 function in the scleral lineage.
Zebrafish alx1 and alx3 function redundantly during ocular morphogenesis
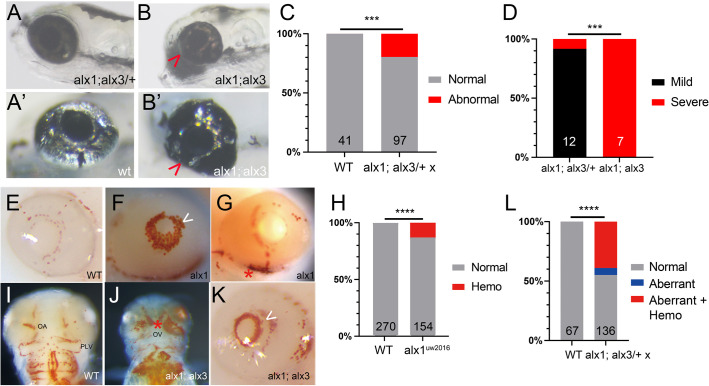
We next asked if alx3 function compensated for the loss of alx1 during ocular morphogenesis. Embryos obtained from a cross between alx1;alx3/+ parents exhibited aberrantly shaped (elongated) eye capsules covered by abnormally patchy iridophores at 5 dpf (Fig. 3A-C). Genotypic analysis confirmed this phenotype, which we termed severe, to be strongly linked to the alx1;alx3 genotype. We also observed a milder manifestation of this ocular phenotype in alx1;alx3 larvae (Fig. 3D).
Fig. 3.
Zebrafish alx1 and alx3 function redundantly during ocular morphogenesis. (A,B) Larvae generated from a cross between alx1;alx3/+ parents were scored at 5 dpf for ocular defects. An alx1;alx3/+ larva with normal ocular morphology (A) similar to that of a wildtype embryo (A′). (B) An alx1;alx3 sibling with ocular coloboma and a misshapen eye (severe defect, arrowheads in B,B′). (C) These ocular defects are restricted to progeny of alx1;alx3/+ parents (***P=0.0009, Fisher's exact test, two trials). (D) The severe ocular defect shown in B is strongly associated with the alx1;alx3 genotype, while the majority of alx1;alx3/+ embryos present with a milder misshapen eye phenotype (not shown) (***P=0.0002 using Fisher's exact test, two trials). (E-L) Embryos derived from wildtype or alx1;alx3/+ parents were stained with o-dianisidine at 2-3 dpf to label hemoglobin in red blood cells. Normal hemoglobin distribution in the eye of a wildtype embryo (E) versus ocular hemorrhage (arrowhead in F) and periocular hemorrhage (asterisk in G) in alx1 mutants. (H) Hemorrhaging is restricted to a subset of alx1 mutants (****P<0.0001, Fisher's exact test, five trials). Normal hemoglobin distribution in the pharyngeal region of a wildtype embryo (I) versus evidence of hemorrhaging in an alx1;alx3 embryo (J). Intraocular and periocular hemorrhages (asterisk) near the optic artery and optic vein in an alx1;alx3 embryo (K). (L) Hemorrhaging is strongly associated with eye defects in alx1;alx3/+ and alx1;alx3 embryos (****P<0.0001, Chi-square test, two trials). Embryos are shown in lateral views, anterior to the left except in I and J, which are ventral views, anterior to the top. OA, optic artery; OV, optic vein; PLV, palatocerebral vein.
Unexpectedly, we also observed sporadic intraocular and periocular hemorrhage in living alx1 and alx1;alx3 embryos at 2-3 dpf. We employed o-dianisidine, a histological stain for erythrocytes, to visualize the aberrantly pooled erythrocytes and to delineate the vascular network. At this stage in development, ocular blood supply is carried by a branched hyaloid network that starts forming around the lens at 32 hpf (Hartsock et al., 2014). In wildtype embryos, erythrocytes were distributed in a pattern that resembled normal hyaloid vasculature (Fig. 3E) (Detrich et al., 1995). This was also true for most alx1 mutants, with a minority exhibiting patches of erythrocytes suggestive of hyaloid hemorrhaging (Fig. 3F-H). In alx1;alx3 homozygotes, o-dianisidine staining revealed extensive and penetrant ocular and periocular hemorrhage (Fig. 3I-K). Ocular hemorrhage was present in ∼50% of alx1;alx3/+ incross progeny and was strongly associated with ocular dysmorphology (Fig. 3L). Thus, hyaloid hemorrhage is a highly penetrant phenotype linked to alx1;alx3 homozygosity. Notably, this defect emerges contemporaneously with the loss of col2a1a expression in ANC precursors and in the sclera (see Fig. 2L, asterisk).
To examine hyaloid vasculature directly, we took advantage of endogenous alkaline phosphatase activity in the vascular endothelium surrounding the lens (Alvarez et al., 2007). Larvae derived from an alx1;alx3/+ incross were sorted by ocular morphology at 5 dpf and stained for alkaline phosphatase. Examination of dissected lenses revealed a range of vascular defects, from mild disorganization to complete absence (Fig. S2). Surprisingly, severity of vascular defects did not correlate with presence or severity of ocular dysmorphology. Together, these data indicate a novel function for the alx family in the developing hyaloid vasculature.
alx1 and alx3 are largely dispensable for early retinal development
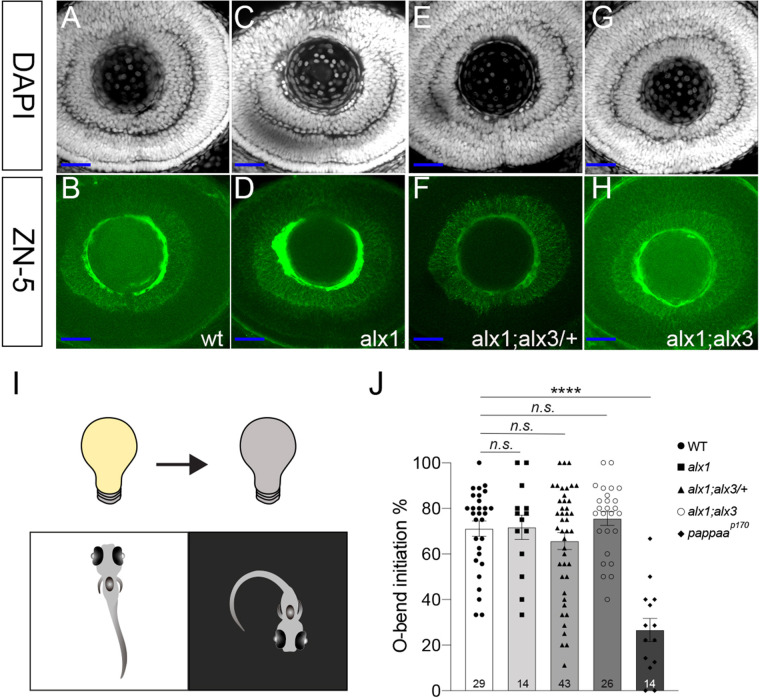
Having documented ocular defects in supporting ocular tissues, we asked if retinal development depended on alx1 and alx3 function. alx1;alx3/+ incross progeny were fixed at 2 dpf and immunostained with an anti-zn5/alcama antibody, which labeled retinal ganglion cell (RGC) neurons, and counter-stained with the nuclear stain DAPI to reveal retinal layers. Confocal imaging revealed a normal pattern of RGCs and correct overall layering in the retinae of alx1, alx1;alx3/+, and alx1;alx3 larvae (Fig. 4A-D).
Fig. 4.
alx1 and alx3 functions are dispensable for retinal ganglion cell differentiation and retinal function in zebrafish larvae. Embryos derived from wildtype or alx1;alx3/+ incrosses were fixed at 2 dpf and stained with zn5, a retinal ganglion cell (RGC) marker, and DAPI, a nuclear co-stain (A-H). (A,B) A representative wildtype eye shows strong zn5-labeled RGCs and a characteristic layered nuclear organization (of four embryos). (C,D) Normal RGC and nuclear patterns in an alx1 mutant (of two embryos). (E,F) Normal RGC and nuclear patterns in a representative alx1;alx3/+ retina (of four embryos). (G,H) Normal RGC and nuclear patterns in a representative alx1;alx3 retina (of two embryos). Left eyes are shown in lateral views, anterior to the left. Scale bar, 25 µm. (I) Diagram summarizing the dark flash assay, with a lights-off stimulus eliciting O-bend turn responses in 5 dpf larval zebrafish. (J) Mean frequency of O-bend initiations to a series of 10 dark flash stimuli was measured in larvae derived from wildtype or alx1;alx3/+ crosses, and larvae were genotyped post-hoc. n.s.: P>0.9999 versus wildtype control; ****P<0.0001 versus wildtype control. ANOVA with Kruskal–Wallis test. Error bars indicate s.e.m.
Next, we tested visually triggered behaviors to assess retinal function in alx1;alx3 larvae at 5 dpf. When presented with an abrupt absence of light (a dark flash), larval zebrafish execute a turning behavior termed the ‘O-bend’, in which they turn their bodies ∼180 degrees (Burgess and Granato, 2007) (Fig. 4I). We exposed 5 dpf larvae to 10 dark flashes and evaluated the probability of O-bend initiation in response to the abrupt absence of light. We found that alx1, alx1;alx3, and alx1;alx3/+ larvae initiated the O-bend at the same frequency as the wildtype larvae (Fig. 4J), while visually impaired pappaap170 mutants showed a marked reduction in O-bend initiation compared to wildtypes (Miller et al., 2018). This analysis shows that alx1;alx3 larvae were able to detect and respond to luminescence changes in their visual field, suggesting their retinae are functional at 5 dpf.
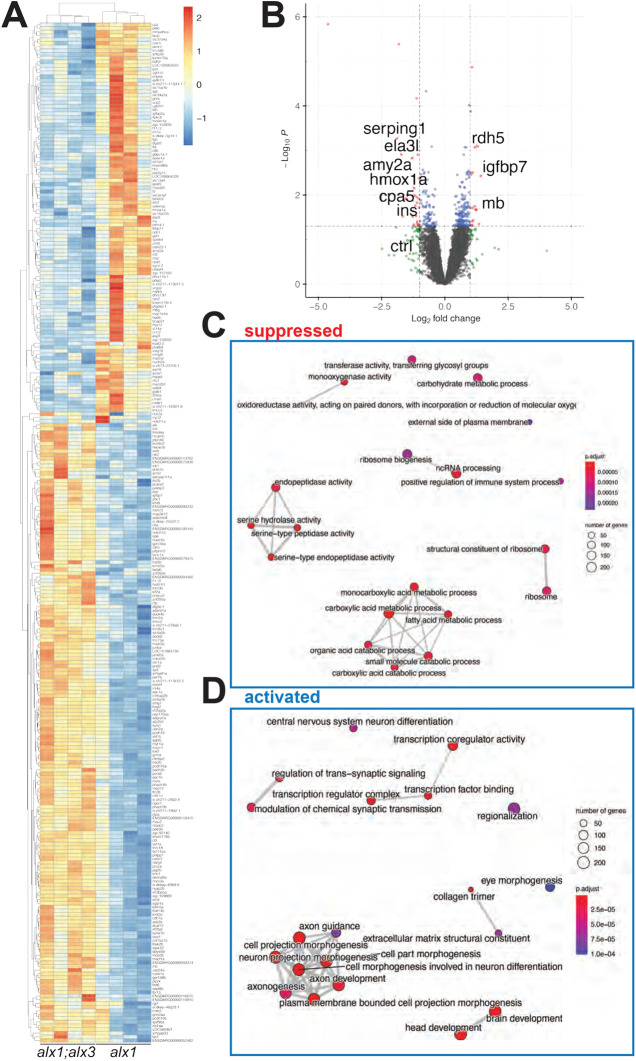
We next set out to identify transcriptional targets that depend on alx1 and alx3 functions in the post-embryonic zebrafish larvae. Embryos were derived from an alx1;alx3/+ incross and RNA was harvested from individual siblings without pooling at 5 dpf, followed by RNA-seq. For each sample, sequence tracks were examined for the presence of the 13 base-pair insertion in the alx3uw2113 allele to deduce the embryonic genotype. 12 samples (four each of alx1, alx1;alx3/+ and alx1;alx3) were selected for pairwise comparisons as detailed in Materials and Methods. This approach identified a compact set of 275 differentially expressed genes (DEGs) between alx1;alx3 and their alx1 siblings (Fig. 5A, Table S1), and 34 genes differentially expressed between alx1;alx3/+ larvae and alx1 siblings (Fig. S3); six of the differentially expressed genes were shared between the two sets. Among DEGs with lineage-restricted expression patterns were several retinal markers that were significantly increased in alx1;alx3 versus alx1 homozygotes at 5 dpf. These differentially expressed genes included cels3, expressed in the amacrine and ganglion cells of the zebrafish retina (Lewis et al., 2011), and rdh5, expressed in retinal pigment epithelium (RPE) (Nadauld et al., 2006). Rdh5 is a retina-specific retinol dehydrogenase required for ventral retinal morphogenesis; its upregulation in alx1;alx3 mutants is consistent with expansion of the RPE. igfpb7, which is highly expressed in vascular endothelium (Abu-Safieh et al., 2011), was also upregulated in alx1;alx3 and in alx1;alx3/+ siblings (Fig. 5B, Fig. S3).
Fig. 5.
RNAseq transcriptome analysis identifies perturbation of ocular development pathways in alx1;alx3 mutants. Larvae derived from alx1uw2016;alx32113/+ parents were lysed individually at 5 dpf for RNA extraction and Illumina high-throughput sequencing. A heat map (A) of significantly dysregulated genes (FDR<0.5) in alx1uw2016;alx3uw2113 double mutants relative to alxuw2016 siblings. A volcano plot (B) of statistical significance against fold-change in alx1uw2016;alx3uw2113 mutants relative to alxuw2016 siblings. Vertical lines indicate −1-fold and +1-fold cut-offs and the horizontal line indicates the FDR=0.05 cut-off. (C,D) The top 20 enriched and top 20 suppressed terms in alx1uw2016;alx3uw2113 double mutants relative to alxuw2016 siblings, identified by GSEA.
The downregulated DEG list was enriched for pancreatic and liver markers, namely, serping1, hmox1a, ela2l, cpa5, ctrl, ela3l, ins and amy2a (Cheng et al., 2006; Holowiecki et al., 2017) suggesting an unexpected role for alx1 and alx3 in these primordia. Two of these genes, serping1 and ela2l, were also downregulated in alx1;alx3/+ versus alx1 siblings (Fig. 5B, Fig. S3).
We next used gene set enrichment analysis (GSEA) (Subramanian et al., 2005) to deeply analyze molecular pathways that require alx1 and alx3 function. This analysis identified several molecular pathways that were suppressed or activated in alx1;alx3 larvae. Ribosome biogenesis was identified as significantly suppressed (Fig. 5C, Fig. S4), while brain and head development, neuronal development and eye morphogenesis were notable among those activated in alx1;alx3 larvae compared to alx1 siblings (Fig. 5D, Fig. S5, Table S2).
Zebrafish alx1 and alx3 function during migration of presumptive anterior neurocranium progenitors
To gain additional insight into the requirement for alx1 and alx3 in the aCNC lineage, we performed in vivo lineage-tracing analysis. To do this efficiently, we adapted CRISPR-driven G0 knockout in zebrafish (Wu et al., 2018) to generate somatic mutations in the alx3 locus in the alx1 homozygous mutant background (alx1;alx3CR, also referred to as ‘crispants’). The alx1;alx3CR larvae exhibited midline clefts, abnormal trabeculae and abnormal anterior neurocranium (ANC) formation (Fig. S6) that resembled the severe defects observed in alx1;alx3 embryos (see Fig. 2A-F). In contrast, alx1 mutants and alx3CR in the wildtype background developed with normal craniofacial cartilages (Fig. S6). Groups of cells in the anterior/frontonasal (aCNC) or maxillary stream (MxCNC), fated to give rise to the ANC (Fig. 6A) (Knight and Schilling, 2006), were labeled by kaede photoconversion in Tg(sox10:kaede) (Fig. 6B,D,F,H) or alx1;alx3CR; Tg(sox10:kaede) embryos (Fig. 6J,L,N,P) at the 20 somite stage and imaged immediately. At 4 dpf, the larvae were imaged again to determine labeled aCNC locations post-migration. Wildtype larvae developed with normal cartilages (labeled in green) and showed normal post-migratory localization of the photo-converted red-kaede-labeled cells (magenta channel) in the median and lateral element of the ANC (Fig. 6C,E,G,I). In alx1;alx3CRs with severely abrogated ANC, red-kaede-labeled cells were found in ectopic locations proximal to the eyes (arrowheads in Fig. 6K,M,O,Q; 17 of 23 total, 2 trials). The median element of the ANC is missing and the lateral element is malformed (Fig. S6). These observations suggest that alx1 and alx3 affect the migration of the frontonasal and maxillary streams of cranial neural crest cells. There also appeared to be fewer crispant cells reaching their destination, consistent with a potential proliferative deficit. Notably, while expression of alx1 and alx3 is primarily NCC-restricted (Dee et al., 2013; Sedykh et al., 2017), these findings do not rule out a non-cell-autonomous function of these transcription factors in tissues adjacent to the migrating neural crest.
Fig. 6.
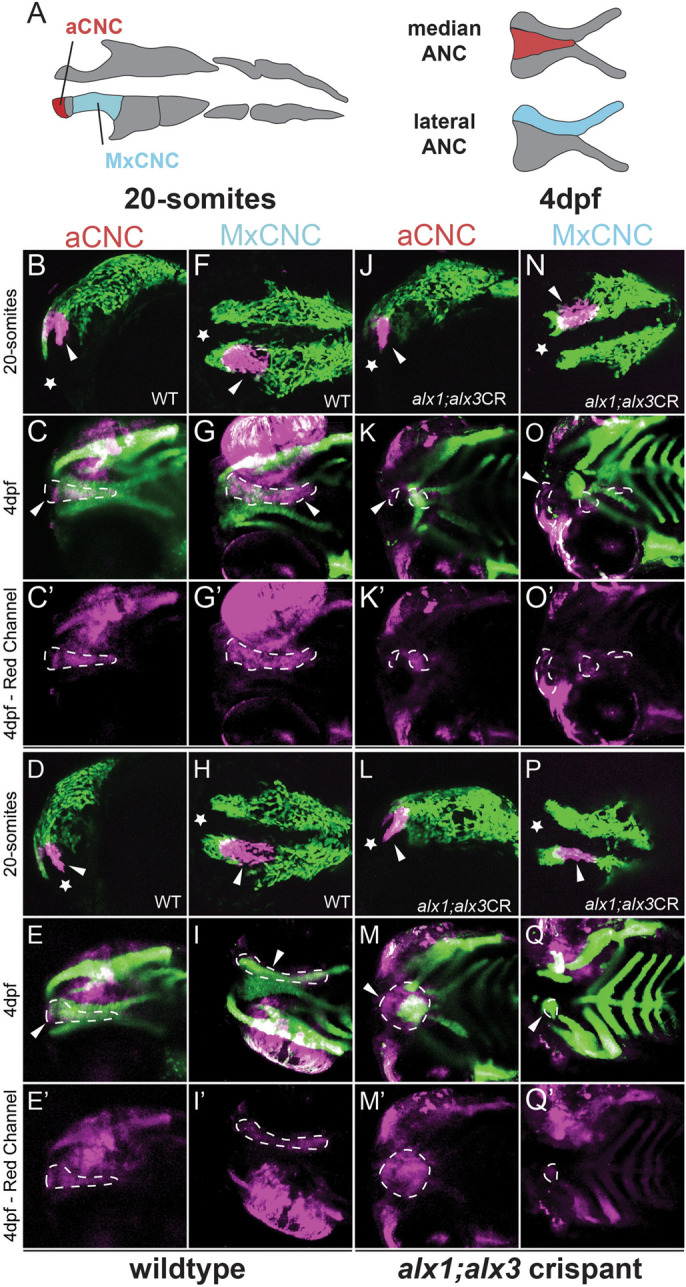
Zebrafish alx1 and alx3 functions are required in migratory anterior cranial neural crest. (A) A schematic fate map of 20-somite-stage cranial neural crest at 4 dpf. The anterior cranial neural crest (aCNC) contributes to the median element of the anterior neurocranium (ANC) while the maxillary stream of migrating CNC (MxCNC) contributes to the maxillary prominence and subsequently coalesce to form the lateral element of the ANC. B-Q: aCNC and MxCNC were labeled by kaede photoconversion (magenta) at 20 somites and imaged immediately, allowed to develop until 4 dpf and imaged again (20-somites images: star sign marks the anterior; 4 dpf images: anterior to the left). alx1-/-;alx3 crispants exhibit a midline cleft (K,M,O,Q) with malformed median and lateral elements. The aCNC and MxCNC migrating streams failed to reach their destinations and ectopically localized along the migratory path (arrowheads in K,M,O,Q). Note that ocular fluorescence is due to the presence of iridophores and retinal pigment epithelium in the eye, not to kaede fluorescence at 4 dpf. Arrowheads point to photoconverted cells and their progeny. Dotted line outlines the shape of the photoconverted cells at 4 dpf. Scale bar: 100 µm.
Transcriptomic analysis identifies candidate targets of Alx1 and Alx3 in the developing embryo
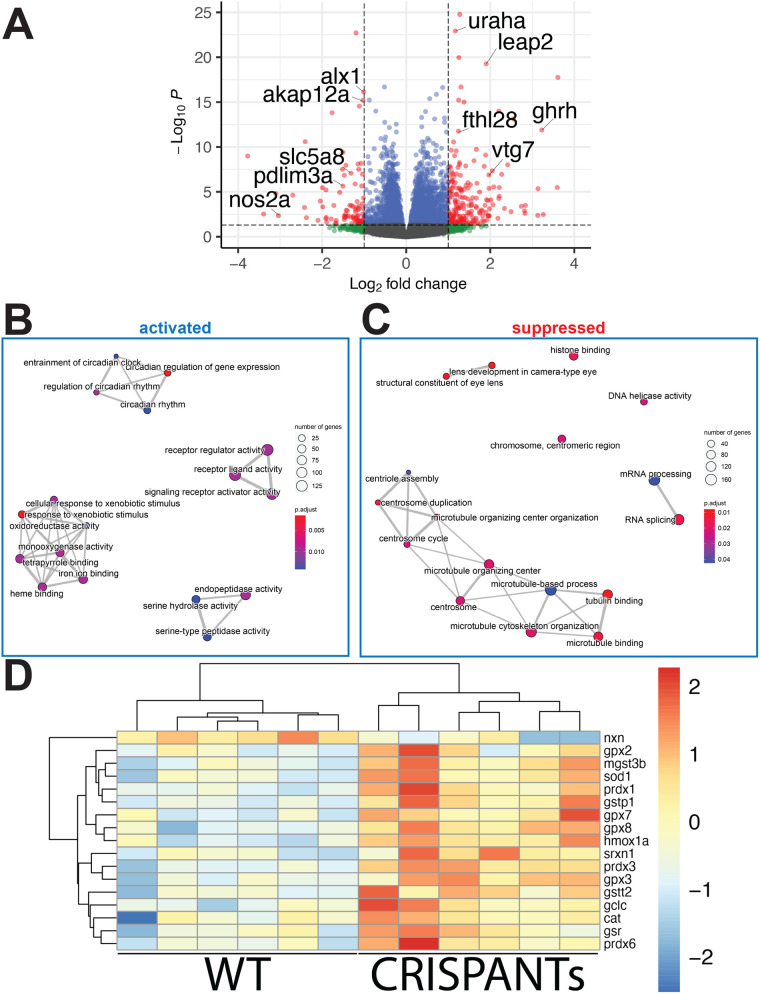
Having demonstrated essential roles for zebrafish alx1 and alx3 in the forming frontonasal cartilage, anterior ocular segment and ocular vasculature, we sought to address the underlying molecular mechanisms through transcriptomic analysis. We surveyed transcriptome changes that result from loss of alx1 and alx3 at 3 dpf, when ocular and ANC defects in mutant embryos become evident. alx1;alx3CR embryos were generated as detailed above, and age-matched wildtype controls were subjected to RNA isolation and high-throughput sequencing. Bioinformatic analysis identified a large set of differentially expressed genes (4426 genes with adjusted P-value <0.05), which included 207 strongly upregulated (Log2 fold change >1) and 93 downregulated (Log2 fold change <−1) genes (Fig. 7A, Table S3).
Fig. 7.
RNAseq transcriptome analysis identifies a role for alx1 and alx3 in oxidative stress response regulation. alx1uw2016;Tg(sox10:kaede) embryos were injected with alx3 CRISPR RNPs to induce somatic mutations at the alx3 locus to generate alx1;alx3CR embryos. At 3 dpf,alx1;alx3CR embryos with characteristic craniofacial defects and age-matched Tg(sox10:kaede) embryos were lysed. A total of six replicate groups of 20 embryos from each genotype were collected in two independent experiments for RNA extraction and Illumina high-throughput sequencing. (A) Volcano plot shows the distribution of genes that are differentially expressed in alx1;alx3CR;sox10:GFP embryos relative to sox10:GFP embryos at 3 dpf. Vertical lines indicate −1.5-fold and +1.5-fold change cut-offs, and the horizontal line indicates the FDR=0.05 cutoff. (B,C) molecular pathways that are significantly suppressed (B) or activated (C) in alx1;alx3CR;sox10:GFP embryos were identified using GSEA, with top 20 activated and top 20 suppressed terms shown as a 2D map to emphasize overlaps. (D) Heatmap summary of differentially expressed oxidative stress response genes shows increased expression levels of these genes in alx1;alx3CR;sox10:GFP embryos.
GSEA analysis of this data set showed suppression of lens development, the only pathway relevant to ocular development, in crispants (Fig. 7B, Fig. S7), while a group of pathways that include monooxygenase activity and xenobiotic metabolism was activated in crispants (Fig. 7C, Fig. S8, Table S4). Notably, several of the leading-edge gene sets contain heme oxygenase 1 (hmox1a), also known for its critical functions in the cellular response to oxidative stress in several tissues, including the vascular endothelium (Fuse et al., 2015; Holowiecki et al., 2017). On further examination of the DEG set, a number of key oxidative-stress-response genes (Nakajima et al., 2011), summarized in the heatmap in Fig. 7D, were found to be upregulated in crispants. These data indicate novel requirements for alx1 and alx3 function in the developing lens, and in regulating the oxidative stress response program.
Zebrafish alx1 protects frontonasal and ocular lineages against ethanol toxicity
The aCNC is particularly sensitive to environmental influences and ethanol, a common environmental toxin and teratogen, is known to impact neural-crest-derived lineages through increasing oxidative stress and disrupting ribosome biogenesis (Kiecker, 2016; Lovely, 2020; Smith et al., 2014). We hypothesized that alx1 function protects aCNC lineages against ethanol toxicity and took advantage of the low penetrance of defects in zebrafish alx1 mutants to test this hypothesis. Embryos derived from alx1×alx1/+ crosses were exposed to 0.5% ethanol from 6 hpf until 48 hpf and stained with Alcian Blue at 5 dpf to visualize craniofacial cartilage (Fig. 8). The exposure dose and duration were selected based on a previous study that demonstrated their minimal effects on embryogenesis in wildtype embryos (McCarthy et al., 2013). As expected, a small proportion of untreated alx1 larvae developed with hypoplastic ANC (Fig. 8A-D). Notably, the penetrance of ANC defects increased significantly in ethanol-exposed alx1 larva (Fig. 2D, P=0.024), while wildtype ANC formation was not significantly affected by ethanol exposure (Fig. 2D, P=0.24).
Fig. 8.
Zebrafish alx1 protects craniofacial and ocular lineages against ethanol toxicity. Embryos derived from wildtype, alx1uw2016, or alx1uw2016/+ parents were exposed to 0.5% ethanol from 6 hpf until 48 hpf and examined for craniofacial and ocular abnormalities at 5 dpf. (A-C) Embryos were stained with Alcian Blue to visualize anterior neurocranium (ANC) chondrocytes. Examples of normal ANC morphology (A) in an ethanol-treated wildtype larva, mildly dysmorphic median ANC in an ethanol-treated alx1uw2016 mutant (B), and severely abrogated ANC with missing median element in an ethanol-treated alx1uw2016 mutant. (D) Ethanol exposure does not increase penetrance of craniofacial defects in wildtype (n.s., not significant; P=0.2424) whereas it increases in progeny from alx1×alx1/+ (*P=0.0237). Compared to untreated or treated wildtype, craniofacial defects are more penetrant in untreated or progeny from alx1×alx1/+ (**P=0.0066, ****, P<0.0001), respectively. Fisher's exact test was used to measure statistical significance (three trials for wildtype and two trials for alx1×alx1/+). (E-G) Embryos were exposed to ethanol and examined for ocular anomalies at 5 dpf. (E) A representative 5 dpf wildtype larva with normal ocular morphology. (F) Misshapen eye in a representative alx1 homozygote. (G) Ethanol exposure increases penetrance of ocular abnormalities in wildtype (*P=0.0448) and in alx1uw2016 mutants (***P=0.0005). Compared to wildtype, abnormal ocular phenotype is more penetrant in both untreated alx1 mutants (***P=0.0003) and in ethanol-treated ones (****P<0.0001). Fisher's exact test was used to measure statistical significance (one trial for wildtype and two trials for alx1 mutants). (H-J) Three representative alx1uw2016 adults exposed to 0.5% ethanol from 6 hpf until 48 hpf and raised to adulthood (2 months of age) exhibit unilateral ocular anomalies. (K) Penetrance of ocular defects increased following embryonic exposure to ethanol but did not rise to significance (n.s., P=0.1022. Fisher's exact test, one trial). (A-C) dissected ANC preparations, anterior to the top. (E,F) lateral views, anterior to the left. H and H′ are different sides of the same fish, as are I and I′, and J and J′, shown in lateral view. mANC: median ANC.
alx1uw2016, but not wildtype, embryos exhibited dysmorphic eyes at 5 dpf (Fig. 8E,F). Ethanol exposure increased the penetrance of this phenotype in alx1 larvae, and caused it to emerge in wildtype larvae, albeit at low penetrance (Fig. 8G). To assess the effect of ethanol exposure on post-embryonic ocular development, ethanol-treated and untreated control alx1uw2016 siblings were raised to adulthood and evaluated for ocular defects at 2 months of age. Ocular defects similar to those documented in Fig. 1 were observed in both groups (Fig. 8H-J). Ethanol-treated alx1uw2016 adults exhibited a higher penetrance of ocular defects than wildtype siblings, but this difference did not rise to significance (P=0.10, Fig. 8J). Together, these data support a novel role for alx1 in conferring robustness to ethanol toxicity on both craniofacial and ocular lineages. Collectively, our findings support crucial functions for alx1 and alx3 in the zebrafish ANC and in several ocular lineages and identify a robust set of candidate molecular mechanisms that underlie these important functions.
DISCUSSION
Here, we show a redundant genetic requirement for zebrafish alx1 and alx3 in several midface structures derived from the aCNC: the ANC, the ocular anterior segment, and ocular blood vessels. We identify a protective role for zebrafish alx1 in ocular and facial embryonic lineages against ethanol toxicity, which, together with the evidence from transcriptome analysis of alx1;alx3 mutants, supports a function for alx1 and alx3 in regulating oxidative stress response and ribosome biogenesis. These findings extend and complement our previous human iPSC studies (Pini et al., 2020) and validate the first animal model of ALX-linked FND with ocular involvement, crucial for understanding conserved roles for ALX genes during vertebrate development.
Complex roles of alx genes during development
Loss of alx1 function has distinct consequences in different vertebrate species. Humans homozygous for ALX1 mutations develop with fully penetrant, severe FND and extreme microopthalmia (Uz et al., 2010; Pini et al., 2020), as do alx1-mutant cats (Lyons et al., 2016). In contrast, mouse alx1 mutants exhibit an earlier and more profound deficit, failure of cranial neural tube closure (Zhao et al., 1996), while zebrafish alx1 mutants develop with variably penetrant, mild craniofacial phenotypes (Pini et al., 2020). Thus, loss of ALX1 function always affects midface development in these vertebrates, but with varying severity and penetrance. These differences may reflect evolutionary divergence in functional specializations of individual ALX genes, common after gene duplication. Comparative genomic studies reveal substantial variability among chordates in the number of alx paralogs, ranging from one in hemichordates to four in zebrafish, likely a result of gene duplication events (Khor and Ettensohn, 2020; Lamichhaney et al., 2015; McGonnell et al., 2011). This variability is thought to have driven emergence of skeletogenesis strategies during chordate evolution (Khor and Ettensohn, 2020). Allelic variation at the alx1 locus is associated with diversity in beak morphologies in Darwin's finch species, thus contributing to rapid evolution (Lamichhaney et al., 2015).
Species-specific differences in alx1 gene functions may also reflect divergence in genetic compensation strategies employed by these organisms. In mouse, alx4 compensates for loss of alx3 since alx3;alx4 double mutants develop with severe orofacial clefts and single mutants do not (Beverdam et al., 2001; Lakhwani et al., 2010). In zebrafish, single alx4a and alx4b mutants develop normally, while alx3 mutants exhibit medial ANC defects (Mitchell et al., 2021). Notably, while Mitchell et al. did not observe craniofacial phenotypes in alx1 mutants (Mitchell et al., 2021), we showed that alx1 mutants develop with partially penetrant ANC defects (Pini et al., 2020). This discrepancy may be due to genetic background variation, which implies the existence of genetic modifiers of alx function that will be important to examine in future studies.
Here we show that genetic removal of both alx1 and alx3 functions significantly increases the penetrance and severity of craniofacial defects compared to alx1 (Pini et al., 2020) and to alx3 single mutants (Mitchell et al., 2021). It is important to note that we did not examine the phenotype of alx3 single mutants in our genetic background, leaving open the possibility that alx3 interacts strongly with genetic modifiers and that the deficits we report here are due entirely to alx3 loss-of-function. However, the absence of defects in alx3 crispants (Fig. S6) argues strongly against this possibility. It is more likely that the relative severity of alx1;alx3 mutant phenotypes reflect functional compensation between alx1 and alx3, potentially through the mechanism of transcriptional adaptation triggered by nonsense-mediated RNA decay (El-Brolosy et al., 2019; Jakutis and Stainier, 2021; Kontarakis and Stainier, 2020; Sztal and Stainier, 2020). This mechanism may also explain why ALX3 does not functionally compensate for the loss of ALX1 function in human ALX1-linked FND, since the disease-linked missense mutations in ALX1 are less likely to trigger nonsense-mediated RNA decay. Collectively, the alx gene family offers an excellent paradigm for asking how genetic compensation is employed by different organisms.
How does alx function in the developing eye?
alx genes are robustly expressed in neural-crest-derived periocular mesenchyme (Dee et al., 2013; Sedykh et al., 2017) that gives rise to the supporting tissues of the eye, including the anterior segment and ocular vascular pericytes (Gage et al., 2005; Jo et al., 2013; Johnston et al., 1979; Le Lievre and Le Douarin, 1975; Trost et al., 2013), but not in retinal lineages. This suggests that the retinal deficits in ALX1 homozygotes are secondary to aberrant formation of supporting structures, and our study provides the first experimental support for this hypothesis. Ocular deficits in post-embryonic alx1 and alx1;alx3 zebrafish appear to be restricted to vascular and anterior segment-derived structures, including the lens, while retinal function, assessed by visually-triggered behaviors, is not affected by loss of alx1 and alx3 (Fig. 4). In contrast, human newborns homozygous for the ALX1 loss-of-function allele present with reduced or absent retina (Fig. 1), and morpholino-mediated alx1 knockdown in zebrafish results in retinal malformations and anterior segment defects, which are likely secondary to retinal defects (Dee et al., 2013). Functional compensation by the remaining zebrafish paralogs, alx4a and alx4b, is a plausible explanation for this discrepancy that can be tested by generating triple and quadruple alx mutants.
It is perhaps not surprising that defects in ocular vasculature have not been observed in ALX1-linked FND patients, since hyaloid vasculature is transient in mammals and regresses prior to birth (Selvam et al., 2018). Our demonstration of a novel role for alx function in hyaloid vessel formation suggests the possibility of a transient defect during early ocular vascular development of FND patients, which could in turn impact retinal development by disrupting endothelial-to-retina signaling recently shown to be required for retinal neurogenesis in zebrafish (Dhakal et al., 2021, 2015). Impaired ocular blood flow may also affect retinal development in mammals which, unlike zebrafish, rely on ocular blood flow for early retinal gas exchange (Dhakal et al., 2015; Pelster and Burggren, 1996).
Neural crest that populates the anterior segment is highly heterogenous; its influence on retinal development during embryogenesis is poorly understood and likely to vary across vertebrates (Takamiya et al., 2020; Van Der Meulen et al., 2020; Williams and Bohnsack, 2015, 2020). It is also likely that the ocular anterior segment offers differing degrees of protection to the developing retina in humans versus lower vertebrates. However, congenital anterior segment dysgenesis (ASD) (Evans and Gage, 2005; Hendee et al., 2018; Portal et al., 2019; Reis and Semina, 2011) is associated with postnatal death of retinal ganglion cells, but not with prenatal retinal defects (Mao et al., 2011, 2017; Martino et al., 2016).
alx1 is also expressed in mouse and chick sclera, a largely overlooked ocular lineage that derives from neural crest and forms adjacent to the retinal pigment epithelium (Thompson et al., 2010). The sclera produces cartilage in birds, while mouse embryos initiate the program but stop short of cartilage production in the sclera (Thompson et al., 2010). Notably, microcalcification of the sclerae and cystic lesions in the anterior chamber have been reported in a patient with splice-site mutation in ALX1 (Uz et al., 2010), lending support for the hypothesis that a role for alx genes in these ocular lineages has been conserved and can be effectively modeled in animals.
Fetal alcohol syndrome and ALX function
Fetal alcohol spectrum disorder (FASD) is a common condition estimated to affect 5% of live births in the US. It presents with disabling neurological defects and is frequently associated with facial and ocular deficits that contribute to the heavy health burden of FASD (Brennan and Giles, 2014; Gupta et al., 2016; Lovely et al., 2017; Lovely, 2020). Emerging evidence indicates that fetal genotype strongly influences the outcome of prenatal ethanol exposure, since many children exposed to high levels of ethanol in utero do not develop FASD (Lovely, 2020). Naturally occurring alleles that confer robustness or sensitivity to alcohol toxicity can be identified epidemiologically, but model organism studies are essential for sustained progress toward a complete understanding of gene-environment interactions that contribute to FASD (de la Morena-Barrio et al., 2018; Eberhart and Parnell, 2016; Kaminen-Ahola, 2020; Lovely et al., 2017).
Model organism studies have shown that neural crest-derived lineages are particularly vulnerable to ethanol toxicity (Smith et al., 2014). Neural crest cells give rise to most craniofacial structures, including the face and the anterior segment of the eyes, and indirectly influence formation of the retina and early brain development. In other words, neural crest vulnerability to ethanol toxicity may account for the majority of defects observed in FASD.
Studies in animal models have offered several mechanistic hypotheses for fetal ethanol toxicity: an increase in reactive oxygen species (ROS), resulting in increased oxidative stress (Brocardo et al., 2011; Dong et al., 2010; Lovely et al., 2017; Wang et al., 2015; Wentzel and Eriksson, 2011), disruption of ribosome biogenesis (Garic et al., 2014) interference with retinoic acid signaling (Williams and Bohnsack, 2019) and apoptosis (McCarthy et al., 2013). Here we show increased vulnerability of facial and ocular lineages to ethanol toxicity in the absence of functional alx1 in zebrafish, and a requirement for alx1 and alx3 function in regulating two of the pathways implicated in FASD: ribosome biogenesis (Fig. 5) and oxidative stress response (Fig. 7). Moreover, mouse Alx3 has been shown to regulate oxidative stress response through activating transcription of Foxo1, a key regulator of the antioxidant pathway (Garcia-Sanz et al., 2017). Notably, the penetrance of ocular defects is lower than the penetrance of craniofacial defects in ethanol treated alx1 mutant embryos, consistent with reports that anterior segment lineages are less susceptible to oxidative stress than craniofacial neural crest (Eason et al., 2017). Our study is the first to demonstrate a protective function for alx1 in the zebrafish model. Real-time in vivo imaging with lineage-specific transgenic reporters, together with pharmacological manipulation of the oxidative stress response pathway, will allow us to rigorously test these functions in future studies. Ultimately, animal models studies will be essential for elucidating the complex, poorly understood processes that contribute to frontonasal development (Farlie et al., 2016).
MATERIALS AND METHODS
Zebrafish strains and embryo manipulation
Adult zebrafish were maintained according to established methods (Westerfield, 1993). All experimental protocols using zebrafish, including adult euthanasia, were approved by the University of Wisconsin Animal Care and Use Committee and carried out in accordance with the institutional animal care protocols. Embryos were obtained from natural mating and staged according to Kimmel (Kimmel et al., 1995). The following mutant strains of zebrafish were used: alx1uw2016 (Pini et al., 2020) and alx1uw2016;alx3uw2113/+ generated in the course of this study (see below) and pappaap170 (Wolman et al., 2015). Embryos were exposed to 0.5% ethanol diluted in E3 starting at 6 hpf. At 2 dpf, embryos were transferred to fresh E3 and raised until 5 dpf to score for craniofacial defects, or until 2 months to score for ocular defects.
CRISPR/Cas9 mutagenesis and mutant allele identification
alx1uw2016;alx3uw2113/+ zebrafish were obtained through alx3-targeted CRISPR/Cas9 mutagenesis in the alx1uw2016 mutant background. Design of alx3 CRISPR site 5′-GGAGTCCCCAGTCAAGCCGT-3′ in exon 2 and mutagenesis were carried out as previously described (Sedykh et al., 2016, 2017), and the recovered mutant alleles were initially characterized by sequencing PCR-amplified genomic DNA. To detect the presence of alx1uw2016 and alx3uw2113 mutant alleles, the relevant region was PCR amplified from genomic DNA extracted from adult tail clips or individual embryos, and subjected to PCR with the following primers: alx1 forward: 5′-CGTGACTTACTGCGCTCCTA-3′ (Pini et al., 2020), alx1 reverse: 5′-CGAGTTCGTCGAGGTCTGTT-3′ (Pini et al., 2020), alx3 forward: 5′-CTATCCCGCTCTGGACTCAG-3′, alx3 reverse: 5′-TCCTCCAGTTGAAAGGTGCT-3′. To characterize mutant alleles, alx3 PCR fragments were subcloned via TA cloning into pGEMT-Easy (Promega) and sequenced. To determine pappaap170 genotype, DNA was extracted from individual larvae and subjected to PCR with the following primers: pappaap170 forward: 5′-CACTCTGGAGCCTCCAGCTTGCGGT-3′ and pappaap170 reverse: 5′-TTGCTGACGTTGTGTACG-3′ (Wolman et al., 2015). The PCR product was digested with Mse1 (New England Biolabs), cleaving the mutant allele and producing a 245 bp fragment distinguishable from the 271 bp wildtype allele. Subsequently, metaphor (Lonza) gel electrophoresis was used to genotype individual embryos and adult fish.
Immunohistochemistry
alx1;alx3/+ incross progeny were fixed at 2 dpf in 4% paraformaldehyde (PFA)/PBS overnight. Their trunk/tail portions were reserved for genotyping and the heads were exposed to 1% KOH/6% H2O2 to remove pigmentation and washed with PBST-X (1X PBS with 0.5% TritonX-100), followed by a 5-min proteinase-K (1:5000) treatment and 30-min 4% PFA/PBS post-fix at room temperature. Embryos were blocked with PBSTD-X (1X PBST, 1% DMSO, 10% BSA, 10% goat serum) for 2 h, at room temperature, incubated with mouse-anti-zn5 primary antibody (Zebrafish International Resource Center A28175, 1:100) and then Alexa-488-conjugated goat anti-mouse secondary antibody (Invitrogen AB_10013770, 1:500). Nuclei were labeled with DAPI (Molecular Probes; 1:5000). For imaging, embryos were mounted in VectaShield (Vector laboratories) and imaged on an Olympus IX81 inverted confocal microscope with the Fluoview 1000 confocal package, using a 60x water immersion objective (NA 1.10).
In situ hybridization and Alcian Blue staining
In situ hybridization was carried out as previously described (Gillhouse et al., 2004) using a probe against col2a1a (Yan et al., 1995). After whole-mount in situ hybridization, embryos were mounted in 100% glycerol and imaged on a Leica MZ FLIII stereo microscope equipped with Leica DFC310 FX camera and LAS v4.0 software. For cartilage staining, larvae were fixed at 5 dpf in 4% PFA/PBS overnight and stained with Alcian Blue according to Kimmel et al. (1998). Stained larvae were further dissected using forceps to isolate jaw cartilage and anterior neurocranium, and imaged using a NIKON eclipse E600 microscope equipped with Q Imaging QIclick 1.4MP CCD Monochrome Microscope camera and NIS-elements software.
O-dianisidine staining and alkaline phosphatase staining
Staining erythrocytes with o-dianisidine was carried out as previously described (Paffett-Lugassy and Zon, 2005). To visualize the intraocular red blood cell population, retinal pigment was removed by exposure to 1% KOH/6% H2O2 and embryos were mounted in 100% glycerol and imaged on Leica MZ FLIII stereo microscopes equipped with Leica DFC310 FX camera and LAS v4.0 software. Endogenous alkaline phosphatase activity in the anterior segment and hyaloid vessels was detected as previously described in (Alvarez et al., 2007; Goehlert et al., 1981; Lessell and Kuwabara, 1964; Vorontsova et al., 2019). Briefly, zebrafish larvae were fixed at 5 dpf in 4% PFA/PBS overnight and eyes were pierced with a sharpened tungsten microneedle and forceps prior to staining. After staining, the retina and sclera were removed with tungsten needles to release the lens, and dissected lenses were imaged using NIKON eclipse E600 microscope equipped with Q Imaging QIclick 1.4MP CCD Monochrome Microscope camera and NIS-elements software.
Adult ocular histology
Adult alx1uw2016 zebrafish were euthanized and preserved in 4% paraformaldehyde fixative. Each fish was photographed on both sides to document eye morphology, and the entire body was immersed for 30 min in a solution of 12% hydrochloric acid (Decal II, Surgipath) to decalcify the bone. Transverse sections were made through the head. The resulting head blocks containing the eyes were submitted for paraffin embedding and step sectioned at 100 μm to sample beyond the midway of the globes. Extra slides were saved at each step. The selected sections were stained using standard protocol with hematoxylin and eosin. Rough measurements were made of each globe in the axial plane and in the vertical plane using the histology slide.
Lineage tracing and fluorescent imaging
Embryos derived from alx1uw2016; Tg(sox10:kaede) were injected with ∼2 nl of 3 mM gRNA:Cas9 RNP complex at the 1-cell stage, as described in a previous study (Hoshijima et al., 2019) to generate alx3 crispants. Injected embryos were raised until the 20-somite stage, whereupon they were dechorionated, selected for strong fluorescent expression, and mounted in low melting agarose containing 0.013% tricaine. Crispants were photoconverted using the 405 nm UV laser on a Leica scanning confocal microscope, with regional selectivity accomplished by using region of interest (ROI) features. Selected cells were exposed to the photoconverting laser at full power for 3-5 min, depending on the size of the region, or until red kaede was observed. Crispants were imaged to record initial position of the photoconverted cells and raised until 4 dpf, when they were once again mounted in low melting agarose with tricaine and imaged on the confocal microscope. For fluorescent imaging of the ANC and Meckel's cartilage, both uninjected siblings and crispants at 4 dpf with different genotypes on a Tg(sox10:kaede) background were dissected and flat-mounted for imaging.
RNA-Seq transcriptome analysis
For the alx1;alx3 mutant dataset, embryos produced from a cross between alx1;alx3/+ parents were flash-frozen individually at 5 dpf, and RNA was prepared using the RNeasy Plus Mini Kit (Qiagen 74134) and a protocol adapted from (de Jong et al., 2010). Genomic DNA contamination was removed using the DNase Max Kit (Qiagen 15200-50). cDNA libraries were prepared using the TruSeq stranded mRNA library preparation protocol with poly-A selection and sequenced on the Illumina HiSeq 2500. Prior to analysis, each sample was genotyped from raw sequencing data alignments using the Integrative Genomics Viewer (Robinson et al., 2011).
For the alx1;alx3CR dataset, embryos derived from alx1uw2016;Tg(sox10:kaede) parents were injected with alx3 sgRNA;Cas9 RNP complex, raised until 3 dpf and screened under fluorescence to select those exhibiting malformed ANC. Embryos derived from Tg(sox10:kaede), fertilized within 2 h and raised under the same conditions were used as age-matched controls. Twenty embryos were combined into each biological replicate, and six replicates for each genotype were collected in two independent experiments. RNA was prepared using the RNeasy Plus Mini Kit (Qiagen 74134) and RNA quality was assessed by Agilent High Sensitivity RNA ScreenTape (Agilent, 5067-5580). cDNA libraries were prepared using the NEBNext® Ultra™ II Directional RNA Library kit (NEB, E7765S) with NEBNext® Poly(A) mRNA Magnetic Isolation Module (E7490S) and sequenced (75 bp paired-end) on the Illumina Nextseq 550.
Bioinformatic analysis of transcriptomic data adhere to recommended ENCODE guidelines and best practices for RNA-Seq (Encode Consortium, 2016). For the alx1;alx3-mutant RNA-seq dataset, alignment of adapter-trimmed (Jiang et al., 2014) (Skewer v0.1.123) 2×150 (paired-end; PE) bp strand-specific Illumina reads to the Danio rerio GRCz11genome (assembly accession GCF_000002035.6) was achieved with the Spliced Transcripts Alignment to a Reference (STAR v2.5.3a) software (Dobin et al., 2013), a splice-junction aware aligner. Expression estimation was performed with RSEM v1.3.0 (RNASeq by Expectation Maximization) (Li and Dewey, 2011). To test for differential gene expression among individual group contrasts, expected read counts obtained from RSEM were used as input into Deseq2 (Love et al., 2014). Pairwise contrasts between alx1;alx3 mutants and age-matched alx1uw2016 siblings were conducted to generate differentially expressed gene lists.
For the alx1;alx3CR RNA-seq dataset, two 75 bp (paired-end; PE) strand-specific Illumina Reads were trimmed to remove Illumina adapter sequences and low-quality sequence with Trimmomatic (version 0.36) (Bolger et al., 2014). Reads were aligned to danRer10 using STAR (version 2.7.1a) (Dobin et al., 2013) with custom built index based on Ensembl v91 gene annotations. Gene expression counts were extracted using featureCounts from the subread package (version 2.0) (Liao et al., 2014) using Ensembl v91 gene annotations. To test for differential gene expression among individual group contrasts, read counts obtained were used as input into Deseq2 (Love et al., 2014). Pairwise contrasts between CRISPants and age-matched wildtype were conducted to generate differentially expressed gene lists. Gene set enrichment was then determined by gene set enrichment analysis (GSEA) (Subramanian et al., 2005) using the Gene Ontology database (Ashburner et al., 2000; Gene Ontology Consortium, 2020).
Behavior testing, video recording and analysis
Zebrafish larvae were raised at 29°C in E3, on a 14:10 h light/dark cycle starting at 1 dpf. On the day of behavioral testing (5 dpf), 60 mm-wide Petri dishes with 25 larvae/dish in 10 ml E3, were placed on a white light box (800 µW/cm2) for at least 1 h. Larvae where transferred to a 4×4 grid and illuminated from above with a mounted LED light (MCWHL5 6500K LED, powered by LEDD1B driver, Thorlabs), and from below with an infrared light source (IR Illuminator CM-IR200B, C&M Vision Technologies). In the grid, larvae were acclimated to the illuminated testing stage (85 µW/cm2) for 5 min before exposure to a series of 10 dark flash stimuli. The O-bend behavior was elicited using an automated behavioral platform in which the mounted LED light and the timing of dark flashes could be controlled (Burgess and Granato, 2007; Wolman et al., 2011). Stimuli were presented at 30 s interstimulus intervals, with each stimulus lasting for 1 s. We used Flote to analyze video images responses in an experimenter-independent, automated manner (Burgess and Granato, 2007). Flote tracks the position of individual larvae frame by frame and characterizes locomotor maneuvers according to predefined kinematic parameters that distinguish these maneuvers. After testing in the 4×4 grid, larvae were transferred to a 96-well plate and genotyped for alx1, alx3, or pappaap170 post-hoc.
Statistics
Statistical analyses, including calculation of means, SEM, and ANOVA, Fisher's exact test and the Chi-square test, were performed using GraphPad Prism software (GraphPad Software). P-values below 0.05 were considered statistically significant.
Supplementary Material
Acknowledgements
We are grateful to Daniel North and Jessica Bethany for expert fish husbandry in our aquarium facilities. We thank Mary Halloran for the gift of the zn5 antibody. We are grateful to the University of Wisconsin Biotechnology Center DNA Sequencing facility for providing sequencing facilities and services, to Mark Berres and the University of Wisconsin Bioinformatics Center for help with RNAseq data analysis, and to the Comparative Ocular Pathology Laboratory of Wisconsin (COPLOW) for adult zebrafish histology.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: Y.G.; Methodology: B.Y., P.Y., L.E.B., K.K., Y.G.; Investigation: B.Y., P.Y., N.S., L.E.B., K.K., J.K., R.D., J.V.O., J.C.; Resources: Y.G.; Writing - original draft: B.Y., Y.G.; Writing - review & editing: Y.G., E.C.L.; Supervision: Y.G., E.C.L.; Project administration: Y.G., E.C.L.; Funding acquisition: Y.G., B.Y., E.C.L.
Funding
This research was supported by the National Institutes of Health [R01EY022098 to Y.G., R35GM119465 to J.C., U01DE024443 and PO1GM061354 to E.C.L.], American Heart Association [Pre-doctoral fellowship award to B.Y.], the National Science Foundation [Graduate Research Fellowship to N.J.S], SciMed GRS [N.J.S.] and Shriners Hospital for Children Research Grants [P.Y., J.K., J.C. and E.C.L.]. Open Access funding was provided by the University of Wisconsin-Madison. Deposited in PMC for immediate release.
Data availability
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (GEO; Edgar et al., 2002) and are accessible through GEO Series accession number GSE202238.
References
- Abu-Safieh, L., Abboud, E. B., Alkuraya, H., Shamseldin, H., Al-Enzi, S., Al-Abdi, L., Hashem, M., Colak, D., Jarallah, A., Ahmad, H.et al. (2011). Mutation of IGFBP7 causes upregulation of BRAF/MEK/ERK pathway and familial retinal arterial macroaneurysms. Am. J. Hum. Genet. 89, 313-319. 10.1016/j.ajhg.2011.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez, Y., Cederlund, M. L., Cottell, D. C., Bill, B. R., Ekker, S. C., Torres-Vazquez, J., Weinstein, B. M., Hyde, D. R., Vihtelic, T. S. and Kennedy, B. N. (2007). Genetic determinants of hyaloid and retinal vasculature in zebrafish. BMC Dev. Biol. 7, 114. 10.1186/1471-213X-7-114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., Davis, A. P., Dolinski, K., Dwight, S. S., Eppig, J. T.et al. (2000). Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25-29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beverdam, A., Brouwer, A., Reijnen, M., Korving, J. and Meijlink, F. (2001). Severe nasal clefting and abnormal embryonic apoptosis in Alx3/Alx4 double mutant mice. Development 128, 3975-3986. 10.1242/dev.128.20.3975 [DOI] [PubMed] [Google Scholar]
- Bolger, A. M., Lohse, M. and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114-2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boschen, K. E., Ptacek, T. S., Berginski, M. E., Simon, J. M. and Parnell, S. E. (2021). Transcriptomic analyses of gastrulation-stage mouse embryos with differential susceptibility to alcohol. Dis. Model. Mech. 14, dmm049012. 10.1242/dmm.049012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan, D. and Giles, S. (2014). Ocular involvement in fetal alcohol spectrum disorder: a review. Curr. Pharm. Des. 20, 5377-5387. 10.2174/1381612820666140205144114 [DOI] [PubMed] [Google Scholar]
- Brocardo, P. S., Gil-Mohapel, J. and Christie, B. R. (2011). The role of oxidative stress in fetal alcohol spectrum disorders. Brain Res. Rev. 67, 209-225. 10.1016/j.brainresrev.2011.02.001 [DOI] [PubMed] [Google Scholar]
- Burgess, H. A. and Granato, M. (2007). Modulation of locomotor activity in larval zebrafish during light adaptation. J. Exp. Biol. 210, 2526-2539. 10.1242/jeb.003939 [DOI] [PubMed] [Google Scholar]
- Cavodeassi, F., Creuzet, S. and Etchevers, H. C. (2019). The hedgehog pathway and ocular developmental anomalies. Hum. Genet. 138, 917-936. 10.1007/s00439-018-1918-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, W., Guo, L., Zhang, Z., Soo, H. M., Wen, C., Wu, W. and Peng, J. (2006). HNF factors form a network to regulate liver-enriched genes in zebrafish. Dev. Biol. 294, 482-496. 10.1016/j.ydbio.2006.03.018 [DOI] [PubMed] [Google Scholar]
- Cvekl, A. and Tamm, E. R. (2004). Anterior eye development and ocular mesenchyme: new insights from mouse models and human diseases. BioEssays 26, 374-386. 10.1002/bies.20009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong, M., Rauwerda, H., Bruning, O., Verkooijen, J., Spaink, H. P. and Breit, T. M. (2010). RNA isolation method for single embryo transcriptome analysis in zebrafish. BMC Res. Notes 3, 73. 10.1186/1756-0500-3-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Morena-Barrio, M. E., Ballesta-Martinez, M. J., López-Gálvez, R., Antón, A. I., López-González, V., Martinez-Ribot, L., Padilla, J., Miñano, A., García-Algar, O., Del Campo, M.et al. (2018). Genetic predisposition to fetal alcohol syndrome: association with congenital disorders of N-glycosylation. Pediatr. Res. 83, 119-127. 10.1038/pr.2017.201 [DOI] [PubMed] [Google Scholar]
- Dee, C. T., Szymoniuk, C. R., Mills, P. E. D. and Takahashi, T. (2013). Defective neural crest migration revealed by a Zebrafish model of Alx1-related frontonasal dysplasia. Hum. Mol. Genet. 22, 239-251. 10.1093/hmg/dds423 [DOI] [PubMed] [Google Scholar]
- Detrich, H. W., III, Kieran, M. W., Chan, F. Y., Barone, L. M., Yee, K., Rundstadler, J. A., Pratt, S., Ransom, D. and Zon, L. I. (1995). Intraembryonic hematopoietic cell migration during vertebrate development. Proc. Natl. Acad. Sci. USA 92, 10713-10717. 10.1073/pnas.92.23.10713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhakal, S., Stevens, C. B., Sebbagh, M., Weiss, O., Frey, R. A., Adamson, S., Shelden, E. A., Inbal, A. and Stenkamp, D. L. (2015). Abnormal retinal development in Cloche mutant zebrafish. Dev. Dyn. 244, 1439-1455. 10.1002/dvdy.24322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhakal, S., Rotem-Bamberger, S., Sejd, J. R., Sebbagh, M., Ronin, N., Frey, R. A., Beitsch, M., Batty, M., Taler, K., Blackerby, J. F.et al. (2021). Selective requirements for vascular endothelial cells and circulating factors in the regulation of retinal neurogenesis. Front. Cell Dev. Biol. 9, 628737. 10.3389/fcell.2021.628737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., Batut, P., Chaisson, M. and Gingeras, T. R. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15-21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, J., Sulik, K. K. and Chen, S.-Y. (2010). The role of NOX enzymes in ethanol-induced oxidative stress and apoptosis in mouse embryos. Toxicol. Lett. 193, 94-100. 10.1016/j.toxlet.2009.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eason, J., Williams, A. L., Chawla, B., Apsey, C. and Bohnsack, B. L. (2017). Differences in neural crest sensitivity to ethanol account for the infrequency of anterior segment defects in the eye compared with craniofacial anomalies in a zebrafish model of fetal alcohol syndrome. Birth Defects Res 109, 1212-1227. 10.1002/bdr2.1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhart, J. K. and Parnell, S. E. (2016). The genetics of fetal alcohol spectrum disorders. Alcohol. Clin. Exp. Res. 40, 1154-1165. 10.1111/acer.13066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Brolosy, M. A., Kontarakis, Z., Rossi, A., Kuenne, C., Günther, S., Fukuda, N., Kikhi, K., Boezio, G. L. M., Takacs, C. M., Lai, S.-L.et al. (2019). Genetic compensation triggered by mutant mRNA degradation. Nature 568, 193-197. 10.1038/s41586-019-1064-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encode Consortium. (2016). https://www.encodeproject.org/documents/cede0cbe-d324-4ce7-ace4-f0c3eddf5972/@download/attachment/ENCODE%20Best%20Practices%20for%20RNA_v2.pdf.
- Evans, A. L. and Gage, P. J. (2005). Expression of the homeobox gene Pitx2 in neural crest is required for optic stalk and ocular anterior segment development. Hum. Mol. Genet. 14, 3347-3359. 10.1093/hmg/ddi365 [DOI] [PubMed] [Google Scholar]
- Garcia-Sanz, P., Mirasierra, M., Moratalla, R. and Vallejo, M. (2017). Embryonic defence mechanisms against glucose-dependent oxidative stress require enhanced expression of alx3 to prevent malformations during diabetic pregnancy. Scientific Reports, 7, 389. 10.1038/s41598-017-00334-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farlie, P. G., Baker, N. L., Yap, P. and Tan, T. Y. (2016). Frontonasal dysplasia: towards an understanding of molecular and developmental aetiology. Mol. Syndromol. 7, 312-321. 10.1159/000450533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsthoefel, P. F. (1963). The embryological development of the effects of Strong's Luxoid gene in the mouse. J. Morphol. 113, 427-451. 10.1002/jmor.1051130307 [DOI] [PubMed] [Google Scholar]
- Fuse, Y., Nakajima, H., Nakajima-Takagi, Y., Nakajima, O. and Kobayashi, M. (2015). Heme-mediated inhibition of Bach1 regulates the liver specificity and transience of the Nrf2-dependent induction of zebrafish heme oxygenase 1. Genes Cells 20, 590-600. 10.1111/gtc.12249 [DOI] [PubMed] [Google Scholar]
- Gage, P. J., Rhoades, W., Prucka, S. K. and Hjalt, T. (2005). Fate maps of neural crest and mesoderm in the mammalian eye. Invest. Ophthalmol. Vis. Sci. 46, 4200-4208. 10.1167/iovs.05-0691 [DOI] [PubMed] [Google Scholar]
- Garic, A., Berres, M. E. and Smith, S. M. (2014). High-throughput transcriptome sequencing identifies candidate genetic modifiers of vulnerability to fetal alcohol spectrum disorders. Alcohol. Clin. Exp. Res. 38, 1874-1882. 10.1111/acer.12457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gene Ontology Consortium. (2020). The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res. 49, D325-D334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gestri, G., Link, B. A. and Neuhauss, S. C. F. (2012). The visual system of zebrafish and its use to model human ocular diseases. Dev. Neurobiol. 72, 302-327. 10.1002/dneu.20919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillhouse, M., Wagner Nyholm, M., Hikasa, H., Sokol, S. Y. and Grinblat, Y. (2004). Two Frodo/Dapper homologs are expressed in the developing brain and mesoderm of zebrafish. Dev. Dyn. 230, 403-409. 10.1002/dvdy.20060 [DOI] [PubMed] [Google Scholar]
- Goehlert, U. G., Ng Ying Kin, N. M. K. and Wolfe, L. S. (1981). Biosynthesis of prostacyclin in rat cerebral microvessels and the choroid plexus. J. Neurochem. 36, 1192-1201. 10.1111/j.1471-4159.1981.tb01718.x [DOI] [PubMed] [Google Scholar]
- Gupta, K. K., Gupta, V. K. and Shirasaka, T. (2016). An update on fetal alcohol syndrome-pathogenesis, risks, and treatment. Alcohol. Clin. Exp. Res. 40, 1594-1602. 10.1111/acer.13135 [DOI] [PubMed] [Google Scholar]
- Hartsock, A., Lee, C., Arnold, V. and Gross, J. M. (2014). In vivo analysis of hyaloid vasculature morphogenesis in zebrafish: a role for the lens in maturation and maintenance of the hyaloid. Dev. Biol. 394, 327-339. 10.1016/j.ydbio.2014.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendee, K. E., Sorokina, E. A., Muheisen, S. S., Reis, L. M., Tyler, R. C., Markovic, V., Cuturilo, G., Link, B. A. and Semina, E. V. (2018). PITX2 deficiency and associated human disease: insights from the zebrafish model. Hum. Mol. Genet. 27, 1675-1695. 10.1093/hmg/ddy074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holowiecki, A., O'Shields, B. and Jenny, M. J. (2017). Spatiotemporal expression and transcriptional regulation of heme oxygenase and biliverdin reductase genes in zebrafish (Danio rerio) suggest novel roles during early developmental periods of heightened oxidative stress. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 191, 138-151. 10.1016/j.cbpc.2016.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshijima, K., Jurynec, M. J., Klatt Shaw, D., Jacobi, A. M., Behlke, M. A. and Grunwald, D. J. (2019). Highly efficient CRISPR-Cas9-based methods for generating deletion mutations and F0 embryos that lack gene function in zebrafish. Dev. Cell 51, 645-657.e644. 10.1016/j.devcel.2019.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakutis, G. and Stainier, D. Y. R. (2021). Genotype-phenotype relationships in the context of transcriptional adaptation and genetic robustness. Annu. Rev. Genet. 55, 71-91. 10.1146/annurev-genet-071719-020342 [DOI] [PubMed] [Google Scholar]
- Jiang, H., Lei, R., Ding, S.-W. and Zhu, S. (2014). Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics 15, 182. 10.1186/1471-2105-15-182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo, D. H., Kim, J. H., Heo, J.-I., Kim, J. H. and Cho, C.-H. (2013). Interaction between pericytes and endothelial cells leads to formation of tight junction in hyaloid vessels. Mol. Cells 36, 465-471. 10.1007/s10059-013-0228-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston, M. C., Noden, D. M., Hazelton, R. D., Coulombre, J. L. and Coulombre, A. J. (1979). Origins of avian ocular and periocular tissues. Exp. Eye Res. 29, 27-43. 10.1016/0014-4835(79)90164-7 [DOI] [PubMed] [Google Scholar]
- Kague, E., Gallagher, M., Burke, S., Parsons, M., Franz-Odendaal, T. and Fisher, S. (2012). Skeletogenic fate of zebrafish cranial and trunk neural crest. PLoS ONE 7, e47394. 10.1371/journal.pone.0047394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminen-Ahola, N. (2020). Fetal alcohol spectrum disorders: genetic and epigenetic mechanisms. Prenat. Diagn. 40, 1185-1192. 10.1002/pd.5731 [DOI] [PubMed] [Google Scholar]
- Khor, J. M. and Ettensohn, C. A. (2020). Transcription factors of the Alx Family: evolutionarily conserved regulators of deuterostome skeletogenesis. Front. Genet. 11, 569314. 10.3389/fgene.2020.569314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiecker, C. (2016). The chick embryo as a model for the effects of prenatal exposure to alcohol on craniofacial development. Dev. Biol. 415, 314-325. 10.1016/j.ydbio.2016.01.007 [DOI] [PubMed] [Google Scholar]
- Kimmel, C. B., Ballard, W. W., Kimmel, S. R., Ullmann, B. and Schilling, T. F. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253-310. 10.1002/aja.1002030302 [DOI] [PubMed] [Google Scholar]
- Kimmel, C. B., Miller, C. T., Kruze, G., Ullmann, B., BreMiller, R. A., Larison, K. D. and Snyder, H. C. (1998). The shaping of pharyngeal cartilages during early development of the zebrafish. Dev. Biol. 203, 245-263. 10.1006/dbio.1998.9016 [DOI] [PubMed] [Google Scholar]
- Kish, P. E., Bohnsack, B. L., Gallina, D., Kasprick, D. S. and Kahana, A. (2011). The eye as an organizer of craniofacial development. Genesis 49, 222-230. 10.1002/dvg.20716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight, R. D. and Schilling, T. F. (2006). Cranial neural crest and development of the head skeleton. Adv. Exp. Med. Biol. 589, 120-133. 10.1007/978-0-387-46954-6_7 [DOI] [PubMed] [Google Scholar]
- Kontarakis, Z. and Stainier, D. Y. R. (2020). Genetics in light of transcriptional adaptation. Trends Genet. 36, 926-935. 10.1016/j.tig.2020.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuratani, S. (2018). The neural crest and origin of the neurocranium in vertebrates. Genesis 56, e23213. 10.1002/dvg.23213 [DOI] [PubMed] [Google Scholar]
- Lakhwani, S., Garcia-Sanz, P. and Vallejo, M. (2010). Alx3-deficient mice exhibit folic acid-resistant craniofacial midline and neural tube closure defects. Dev. Biol. 344, 869-880. 10.1016/j.ydbio.2010.06.002 [DOI] [PubMed] [Google Scholar]
- Lamichhaney, S., Berglund, J., Almén, M. S., Maqbool, K., Grabherr, M., Martinez-Barrio, A., Promerová, M., Rubin, C.-J., Wang, C., Zamani, N.et al. (2015). Evolution of Darwin's finches and their beaks revealed by genome sequencing. Nature 518, 371-375. 10.1038/nature14181 [DOI] [PubMed] [Google Scholar]
- Le Lievre, C. S. and Le Douarin, N. M. (1975). Mesenchymal derivatives of the neural crest: analysis of chimaeric quail and chick embryos. J. Embryol. Exp. Morphol. 34, 125-154. 10.1242/dev.34.1.125 [DOI] [PubMed] [Google Scholar]
- Lessell, S. and Kuwabara, T. (1964). Phosphatase Histochemistry of the Eye. Arch. Ophthalmol. 71, 851-860. 10.1001/archopht.1964.00970010867015 [DOI] [PubMed] [Google Scholar]
- Lewis, A., Wilson, N., Stearns, G., Johnson, N., Nelson, R. and Brockerhoff, S. E. (2011). Celsr3 is required for normal development of GABA circuits in the inner retina. PLoS Genet. 7, e1002239. 10.1371/journal.pgen.1002239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B. and Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323. 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, Y., Smyth, G. K. and Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923-930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Love, M. I., Huber, W. and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovely, C. B. (2020). Animal models of gene-alcohol interactions. Birth Defects Res 112, 367-379. 10.1002/bdr2.1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovely, C., Rampersad, M., Fernandes, Y. and Eberhart, J. (2017). Gene-environment interactions in development and disease. Wiley Interdiscipl. Rev. Dev. Biol. 6, e247. 10.1002/wdev.247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons, L. A., Erdman, C. A., Grahn, R. A., Hamilton, M. J., Carter, M. J., Helps, C. R., Alhaddad, H. and Gandolfi, B. (2016). Aristaless-Like Homeobox protein 1 (ALX1) variant associated with craniofacial structure and frontonasal dysplasia in Burmese cats. Dev. Biol. 409, 451-458. 10.1016/j.ydbio.2015.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J. and Lwigale, P. (2019). Transformation of the transcriptomic profile of mouse periocular mesenchyme during formation of the embryonic cornea. Invest. Ophthalmol. Vis. Sci. 60, 661-676. 10.1167/iovs.18-26018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, M., Hedberg-Buenz, A., Koehn, D., John, S. W. M. and Anderson, M. G. (2011). Anterior segment dysgenesis and early-onset glaucoma in nee mice with mutation of Sh3pxd2b. Invest. Ophthalmol. Vis. Sci. 52, 2679-2688. 10.1167/iovs.10-5993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, M., Kiss, M., Ou, Y. and Gould, D. B. (2017). Genetic dissection of anterior segment dysgenesis caused by a Col4a1 mutation in mouse. Dis. Model. Mech. 10, 475-485. 10.1242/dmm.027888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino, V. B., Sabljic, T., Deschamps, P., Green, R. M., Akula, M., Peacock, E., Ball, A., Williams, T. and West-Mays, J. A. (2016). Conditional deletion of AP-2beta in mouse cranial neural crest results in anterior segment dysgenesis and early-onset glaucoma. Dis. Model. Mech. 9, 849-861. 10.1242/dmm.025262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy, N., Wetherill, L., Lovely, C. B., Swartz, M. E., Foroud, T. M. and Eberhart, J. K. (2013). Pdgfra protects against ethanol-induced craniofacial defects in a zebrafish model of FASD. Development 140, 3254-3265. 10.1242/dev.094938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGonnell, I. M., Graham, A., Richardson, J., Fish, J. L., Depew, M. J., Dee, C. T., Holland, P. W. H. and Takahashi, T. (2011). Evolution of the Alx homeobox gene family: parallel retention and independent loss of the vertebrate Alx3 gene. Evol. Dev. 13, 343-351. 10.1111/j.1525-142X.2011.00489.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, A. H., Howe, H. B., Krause, B. M., Friedle, S. A., Banks, M. I., Perkins, B. D. and Wolman, M. A. (2018). Pregnancy-associated plasma protein-aa regulates photoreceptor synaptic development to mediate visually guided behavior. J. Neurosci. 38, 5220-5236. 10.1523/JNEUROSCI.0061-18.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, J. M., Sucharov, J., Pulvino, A. T., Brooks, E. P., Gillen, A. E. and Nichols, J. T. (2021). The alx3 gene shapes the zebrafish neurocranium by regulating frontonasal neural crest cell differentiation timing. Development 148, dev197483. 10.1242/dev.197483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadauld, L. D., Chidester, S., Shelton, D. N., Rai, K., Broadbent, T., Sandoval, I. T., Peterson, P. W., Manos, E. J., Ireland, C. M., Yost, H. J.et al. (2006). Dual roles for adenomatous polyposis coli in regulating retinoic acid biosynthesis and Wnt during ocular development. Proc. Natl. Acad. Sci. USA 103, 13409-13414. 10.1073/pnas.0601634103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima, H., Nakajima-Takagi, Y., Tsujita, T., Akiyama, S.-I., Wakasa, T., Mukaigasa, K., Kaneko, H., Tamaru, Y., Yamamoto, M. and Kobayashi, M. (2011). Tissue-restricted expression of Nrf2 and its target genes in zebrafish with gene-specific variations in the induction profiles. PLoS ONE 6, e26884. 10.1371/journal.pone.0026884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paffett-Lugassy, N. N. and Zon, L. I. (2005). Analysis of hematopoietic development in the zebrafish. Methods Mol. Med. 105, 171-198. 10.1385/1-59259-826-9:171 [DOI] [PubMed] [Google Scholar]
- Pelster, B. and Burggren, W. W. (1996). Disruption of hemoglobin oxygen transport does not impact oxygen-dependent physiological processes in developing embryos of zebra fish (Danio rerio). Circ. Res. 79, 358-362. 10.1161/01.RES.79.2.358 [DOI] [PubMed] [Google Scholar]
- Pini, J., Kueper, J., Hu, Y. D., Kawasaki, K., Yeung, P., Tsimbal, C., Yoon, B., Carmichael, N., Maas, R. L., Cotney, J.et al. (2020). ALX1-related frontonasal dysplasia results from defective neural crest cell development and migration. EMBO Mol. Med. 12, e12013. 10.1101/2020.06.12.148262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portal, C., Rompolas, P., Lwigale, P. and Iomini, C. (2019). Primary cilia deficiency in neural crest cells models anterior segment dysgenesis in mouse. eLife 8, e52423. 10.7554/eLife.52423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu, S., Tucker, S. C., Zhao, Q., deCrombrugghe, B. and Wisdom, R. (1999). Physical and genetic interactions between Alx4 and Cart1. Development 126, 359-369. 10.1242/dev.126.2.359 [DOI] [PubMed] [Google Scholar]
- Reis, L. M. and Semina, E. V. (2011). Genetics of anterior segment dysgenesis disorders. Curr. Opin Ophthalmol. 22, 314-324. 10.1097/ICU.0b013e328349412b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, J. T., Thorvaldsdottir, H., Winckler, W., Guttman, M., Lander, E. S., Getz, G. and Mesirov, J. P. (2011). Integrative genomics viewer. Nat. Biotechnol. 29, 24-26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedykh, I., TeSlaa, J. J., Tatarsky, R. L., Keller, A. N., Toops, K. A., Lakkaraju, A., Nyholm, M. K., Wolman, M. A. and Grinblat, Y. (2016). Novel roles for the radial spoke head protein 9 in neural and neurosensory cilia. Sci. Rep. 6, 34437. 10.1038/srep34437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedykh, I., Yoon, B., Roberson, L., Moskvin, O., Dewey, C. N. and Grinblat, Y. (2017). Zebrafish zic2 controls formation of periocular neural crest and choroid fissure morphogenesis. Dev. Biol. 429, 92-104. 10.1016/j.ydbio.2017.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvam, S., Kumar, T. and Fruttiger, M. (2018). Retinal vasculature development in health and disease. Prog. Retin. Eye Res. 63, 1-19. 10.1016/j.preteyeres.2017.11.001 [DOI] [PubMed] [Google Scholar]
- Serbedzija, G. N., Bronner-Fraser, M. and Fraser, S. E. (1992). Vital dye analysis of cranial neural crest cell migration in the mouse embryo. Development 116, 297-307. 10.1242/dev.116.2.297 [DOI] [PubMed] [Google Scholar]
- Smith, S. M., Garic, A., Flentke, G. R. and Berres, M. E. (2014). Neural crest development in fetal alcohol syndrome. Birth Defects Res. C Embryo Today 102, 210-220. 10.1002/bdrc.21078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian, A., Tamayo, P., Mootha, V. K., Mukherjee, S., Ebert, B. L., Gillette, M. A., Paulovich, A., Pomeroy, S. L., Golub, T. R., Lander, E. S.et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545-15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztal, T. E. and Stainier, D. Y. R. (2020). Transcriptional adaptation: a mechanism underlying genetic robustness. Development 147, dev186452. 10.1242/dev.186452 [DOI] [PubMed] [Google Scholar]
- Takamiya, M., Stegmaier, J., Kobitski, A. Y., Schott, B., Weger, B. D., Margariti, D., Cereceda Delgado, A. R., Gourain, V., Scherr, T., Yang, L.et al. (2020). Pax6 organizes the anterior eye segment by guiding two distinct neural crest waves. PLoS Genet. 16, e1008774. 10.1371/journal.pgen.1008774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Berge, D., Brouwer, A., el Bahi, S., Guénet, J.-L., Robert, B. and Meijlink, F. (1998). Mouse Alx3: an aristaless-like homeobox gene expressed during embryogenesis in ectomesenchyme and lateral plate mesoderm. Dev. Biol. 199, 11-25. 10.1006/dbio.1998.8921 [DOI] [PubMed] [Google Scholar]
- Thompson, H., Griffiths, J. S., Jeffery, G. and McGonnell, I. M. (2010). The retinal pigment epithelium of the eye regulates the development of scleral cartilage. Dev. Biol. 347, 40-52. 10.1016/j.ydbio.2010.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trost, A., Schroedl, F., Lange, S., Rivera, F. J., Tempfer, H., Korntner, S., Stolt, C. C., Wegner, M., Bogner, B., Kaser-Eichberger, A.et al. (2013). Neural crest origin of retinal and choroidal pericytes. Invest. Ophthalmol. Vis. Sci. 54, 7910-7921. 10.1167/iovs.13-12946 [DOI] [PubMed] [Google Scholar]
- Twigg, S. R. F., Versnel, S. L., Nürnberg, G., Lees, M. M., Bhat, M., Hammond, P., Hennekam, R. C. M., Hoogeboom, A. J. M., Hurst, J. A., Johnson, D.et al. (2009). Frontorhiny, a distinctive presentation of frontonasal dysplasia caused by recessive mutations in the ALX3 homeobox gene. Am. J. Hum. Genet. 84, 698-705. 10.1016/j.ajhg.2009.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uz, E., Alanay, Y., Aktas, D., Vargel, I., Gucer, S., Tuncbilek, G., von Eggeling, F., Yilmaz, E., Deren, O., Posorski, N.et al. (2010). Disruption of ALX1 causes extreme microphthalmia and severe facial clefting: expanding the spectrum of autosomal-recessive ALX-related frontonasal dysplasia. Am. J. Hum. Genet. 86, 789-796. 10.1016/j.ajhg.2010.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Meulen, K. L., Vöcking, O., Weaver, M. L., Meshram, N. N. and Famulski, J. K. (2020). Spatiotemporal characterization of anterior segment mesenchyme heterogeneity during zebrafish ocular anterior segment development. Front. Cell Dev. Biol. 8, 379. 10.3389/fcell.2020.00379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorontsova, I., Hall, J. E. and Schilling, T. F. (2019). Assessment of zebrafish lens nucleus localization and sutural integrity. J. Vis. Exp. 147, e59528. 10.3791/59528 [DOI] [PubMed] [Google Scholar]
- Wada, N., Javidan, Y., Nelson, S., Carney, T. J., Kelsh, R. N. and Schilling, T. F. (2005). Hedgehog signaling is required for cranial neural crest morphogenesis and chondrogenesis at the midline in the zebrafish skull. Development 132, 3977-3988. 10.1242/dev.01943 [DOI] [PubMed] [Google Scholar]
- Wang, X.-Y., Li, S., Wang, G., Ma, Z.-L., Chuai, M., Cao, L. and Yang, X. (2015). High glucose environment inhibits cranial neural crest survival by activating excessive autophagy in the chick embryo. Sci. Rep. 5, 18321. 10.1038/srep18321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentzel, P. and Eriksson, U. J. (2011). Altered gene expression in rat cranial neural crest cells exposed to a teratogenic glucose concentration in vitro-paradoxical downregulation of antioxidative defense genes. Birth Defects Res. B Dev. Reprod. Toxicol. 92, 487-497. 10.1002/bdrb.20321 [DOI] [PubMed] [Google Scholar]
- Westerfield, M. (1993). The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Brachydanio rerio). Eugene, OR: M. Westerfield. [Google Scholar]
- Williams, A. L. and Bohnsack, B. L. (2015). Neural crest derivatives in ocular development: discerning the eye of the storm. Birth Defects Res. C Embryo Today 105, 87-95. 10.1002/bdrc.21095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, A. L. and Bohnsack, B. L. (2019). What's retinoic acid got to do with it? Retinoic acid regulation of the neural crest in craniofacial and ocular development. Genesis 57, e23308. 10.1002/dvg.23308 [DOI] [PubMed] [Google Scholar]
- Williams, A. L. and Bohnsack, B. L. (2020). The ocular neural crest: specification, migration, and then what? Front. Cell Dev. Biol. 8, 595896. 10.3389/fcell.2020.595896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolman, M. A., Jain, R. A., Liss, L. and Granato, M. (2011). Chemical modulation of memory formation in larval zebrafish. Proc. Natl. Acad. Sci. USA 108, 15468-15473. 10.1073/pnas.1107156108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolman, M. A., Jain, R. A., Marsden, K. C., Bell, H., Skinner, J., Hayer, K. E., Hogenesch, J. B. and Granato, M. (2015). A genome-wide screen identifies PAPP-AA-mediated IGFR signaling as a novel regulator of habituation learning. Neuron 85, 1200-1211. 10.1016/j.neuron.2015.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, R. S., Lam, I. I., Clay, H., Duong, D. N., Deo, R. C. and Coughlin, S. R. (2018). A rapid method for directed gene knockout for screening in G0 zebrafish. Dev. Cell 46, 112-125.e114. 10.1016/j.devcel.2018.06.003 [DOI] [PubMed] [Google Scholar]
- Yan, Y.-L., Hatta, K., Riggleman, B. and Postlethwait, J. H. (1995). Expression of a type II collagen gene in the zebrafish embryonic axis. Dev. Dyn. 203, 363-376. 10.1002/aja.1002030308 [DOI] [PubMed] [Google Scholar]
- Zhao, Q., Behringer, R. R. and de Crombrugghe, B. (1996). Prenatal folic acid treatment suppresses acrania and meroanencephaly in mice mutant for the Cart1 homeobox gene. Nat. Genet. 13, 275-283. 10.1038/ng0796-275 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.