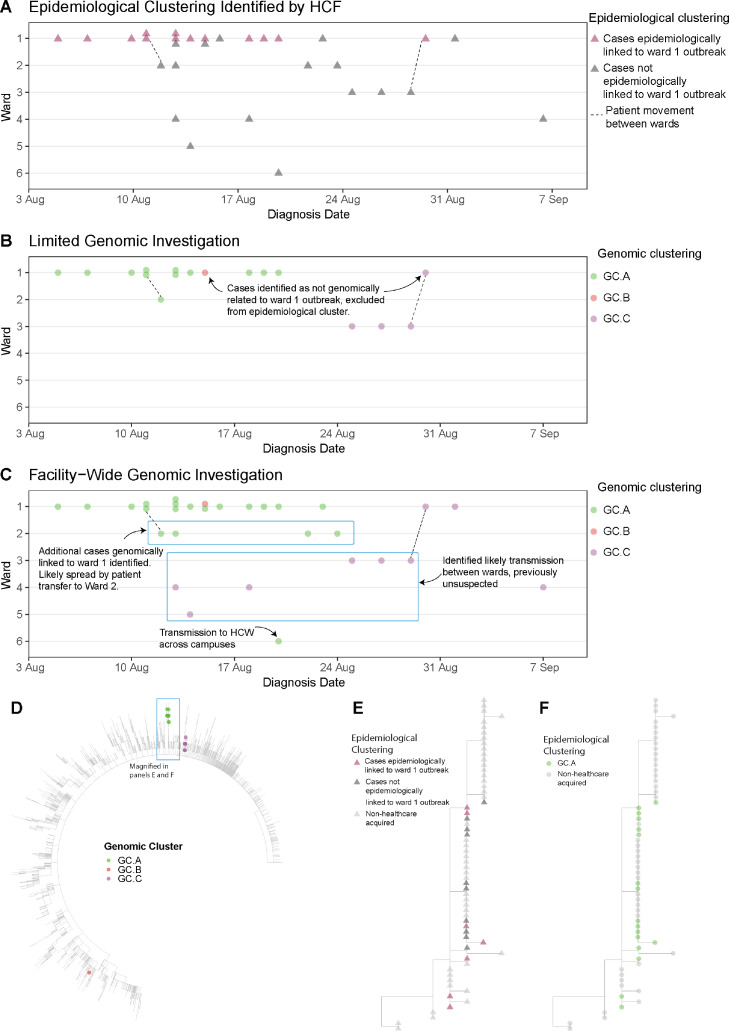
Figure 3.
Comparison of clustering (identifying cases of HCW and patient infections that are likely to be related) at Facility C using three different models: epidemiological clustering (Panel A), limited genomic investigation (cases in a single ward selected by HCF, panel B), and facility-wide genomic infections (Panel C). Each panel shows the distribution of cases (triangles in panel A, circles in panels B and C) across six different wards (Wards 1-6) over a six-week time period in 2020. In panel A, thirteen cases were identified by the HCF as a likely epidemiologic cluster (pink triangles). These cases, with the addition three cases from adjacent ward (Ward 3) were submitted for a limited genomic investigation (Panel B); cases (circles) are coloured by genomic cluster. This showed that most of the cases submitted were part of the same genomic cluster, but two of the Ward 1 cases were not linked (one case from GC.B, and one case from GC.C, which was linked to two other cases on Ward 3). Panel C shows a broader facility-wide genomic investigation that was undertaken to investigate cases on other wards; all HCW and patient cases were included in the facility-wide investigation. This genomic analysis found the main outbreak from Ward 1 was larger than first identified, linking outbreaks in adjacent wards to the Ward 1 outbreak, with cryptic transmission between wards resulting in spread, including transmission to another hospital campus. Unexpected links were also identified for GC.C, with cases spread over four wards. These genomic links were used to direct further investigations to identify causes of transmission and introduce mitigation strategies. Panel D shows a maximum likelihood phylogenetic tree of Facility C cases with Australian SARS-CoV-2 samples from wave 2, July – October 2020. Colour indicates cluster; genomic clustering labels are independent from previously presented analyses (labels simplified for ease of communication). Panel E shows a sub-section of the tree of panel D, with nodes coloured by epidemiologic clustering identified by the HCF, as in panel A; cases thought likely part of epidemiologic cluster (pink triangles), not epidemiologically linked (dark grey triangles) or not associated with the HCF (light grey triangles). Panel F shows same sub-tree as panel E, but coloured by genomic cluster; GC.A indicates the genomic cluster originally identified in ward 1 as in panels B and C; light grey circles indicate samples not associated with the HCF.