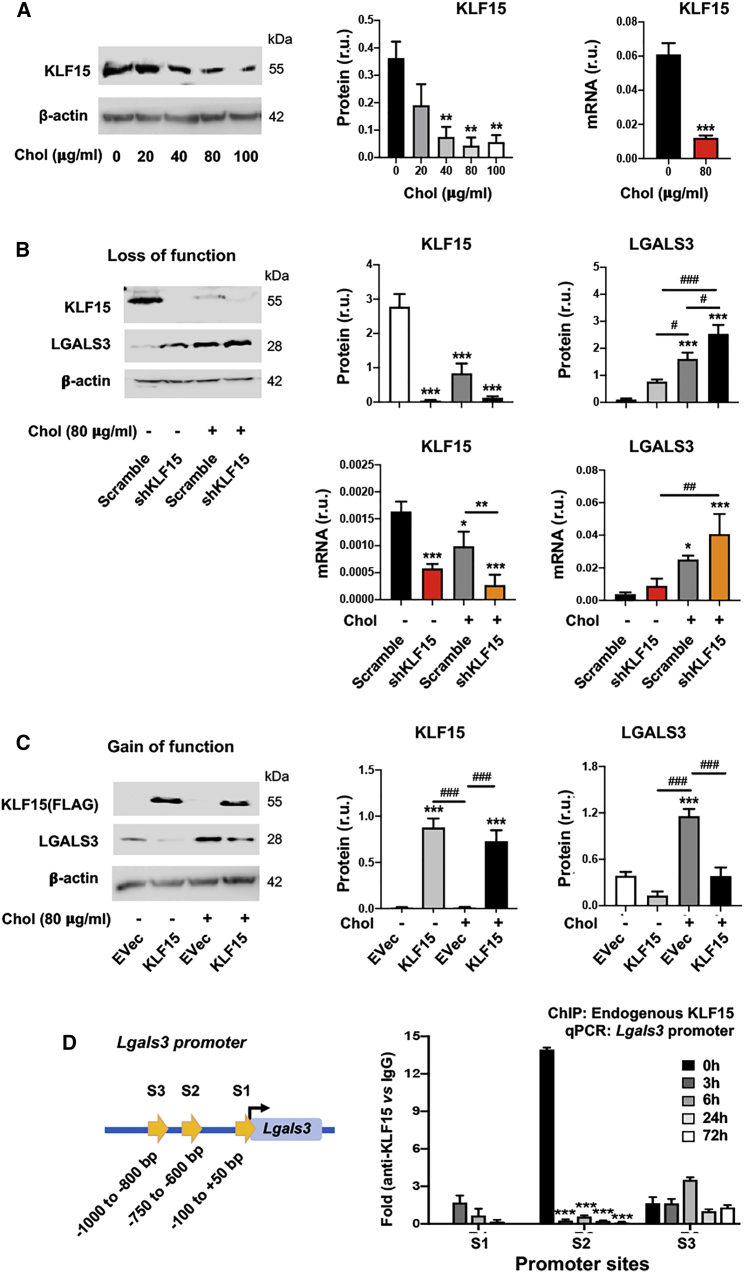
Figure 2.
KLF15 negatively regulates LGALS3 expression in mouse SMCs
Mouse MOVAS cells were used. Cholesterol loading and lentiviral transduction were performed as described in detail for Figure 1. Data quantification for western blots: mean ± SEM, n ≥ 3 independent repeat experiments. Data quantification for quantitative real-time PCR and ChIP-qPCR: mean ± SD, n = 3 replicates; presented is one of two similar experiments. Statistics: ANOVA followed by Tukey test; #p < 0.05, ##p < 0.01, ###p < 0.001 (between paired bars); ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, compared with the basal condition (the first bar in each plot). (A) Down-regulation of KLF15 by increasing concentrations of cholesterol. (B) KLF15 silencing increases LGALS3 protein and mRNA. (C) KLF15 overexpression reduces LGALS3 protein. (D) ChIP-qPCR indicating KLF15 binding at the S2 site of the Lgals3 promoter. S1, S2, and S3 refer to the same sites in Figure 1E. ChIP was performed using an antibody specific for endogenous KLF15, after MOVAS cells were treated for indicated hours.