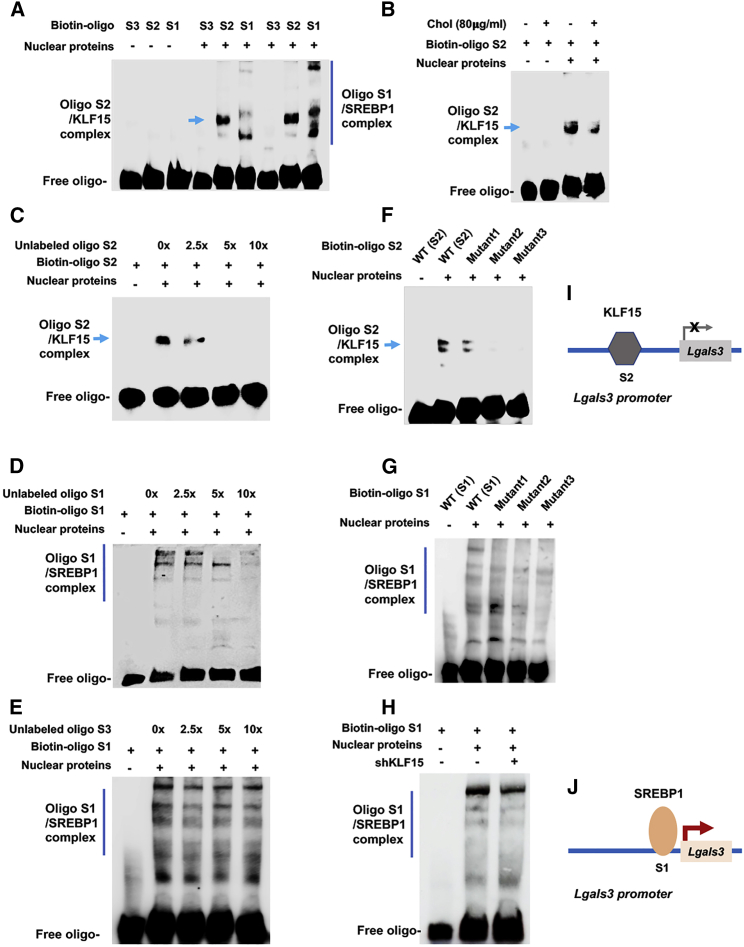
Figure 3.
Electrophoretic mobility shift assays (EMSAs) showing transcription factor/promoter DNA interactions
MOVAS cells were cultured without or with 80 μg/mL cholesterol for 72 h. Nuclear proteins were then extracted and incubated with biotin-labeled oligonucleotides (A and B). Duplicate samples were loaded to the right six lanes in (A). S1, S2, and S3 denote the oligo sequences within the respective S1, S2, and S3 Lgals3 promoter sites (see Figures 1E and 2D). (C–E) A competitor oligo (without biotin label) in 2.5×, 5×, or 10× molar excess was added in nuclear extracts before adding the biotin-oligo of the same sequence. (F and G) A biotin-oligo without (WT control) or with a mutated sequence (mutant) was used. Shown in each blot is one of two to three similar experiments. (H) Experiments were performed in cells transduced with lentivirus expressing scrambled or KLF15-specific shRNA. (I and J) Schematics depict KLF15’s negative effect and SREBP1’s positive effect on Lgals3 transcription, respectively, as suggested by combined results presented in Figures 1 and 2 and this figure.