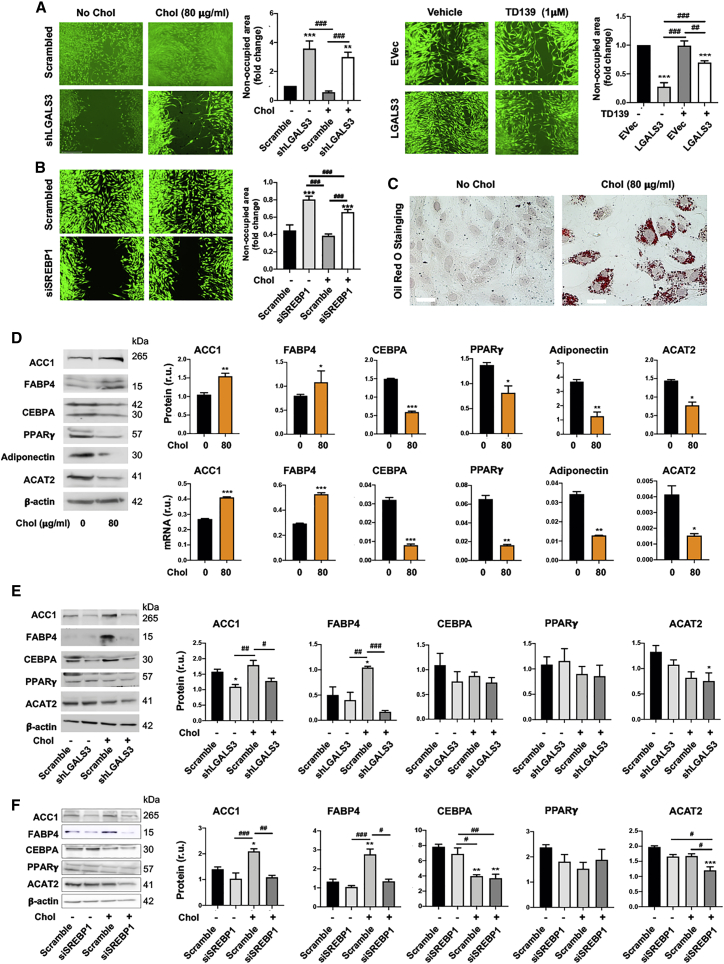
Figure 7.
LGALS3 mediates SMC migration and activates expression of a subset of adipogenic marker genes
Mouse MOVAS cells were used unless otherwise specified (i.e., rat primary aortic SMCs in (B) and (F). Cholesterol loading (80 μg/mL) and lentiviral transduction were performed as described in detail for Figure 1. Data quantification for western blots: mean ± SEM, n ≥ 3 independent repeat experiments. Data quantification for quantitative real-time PCR: mean ± SD, n = 3 replicates; presented is one of two similar experiments. Statistics: ANOVA followed by Tukey test; #p < 0.05, ##p < 0.01, ###p < 0.001 (between paired bars); ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, compared with the basal condition (the first bar in each plot). (A) Effect of LGALS3 silencing or overexpression on MOVAS cell migration. Cells were illuminated with calcein fluorescence. Larger non-occupied scratch wound area indicates less migration. (B) Effect of SREBP1 silencing on rat primary SMC migration. Cells were illuminated with calcein fluorescence. (C) Lipid storage in MOVAS cells. Shown are representative images of oil red O staining. Unstained circles are nuclei. Scale bar: 100 μm. (D) Effect of cholesterol loading (to MOVAS cells) on adipogenic marker expression (protein and mRNA). (E) Effect of LGALS3 silencing on protein levels of adipogenic markers. MOVAS cells were transduced with lentivirus expressing scrambled or LGALS3-specific shRNA. (F) Effect of SREBP1 silencing on protein levels of adipogenic markers. Rat primary SMCs were transfected with scrambled or SREBP1-specific siRNA.