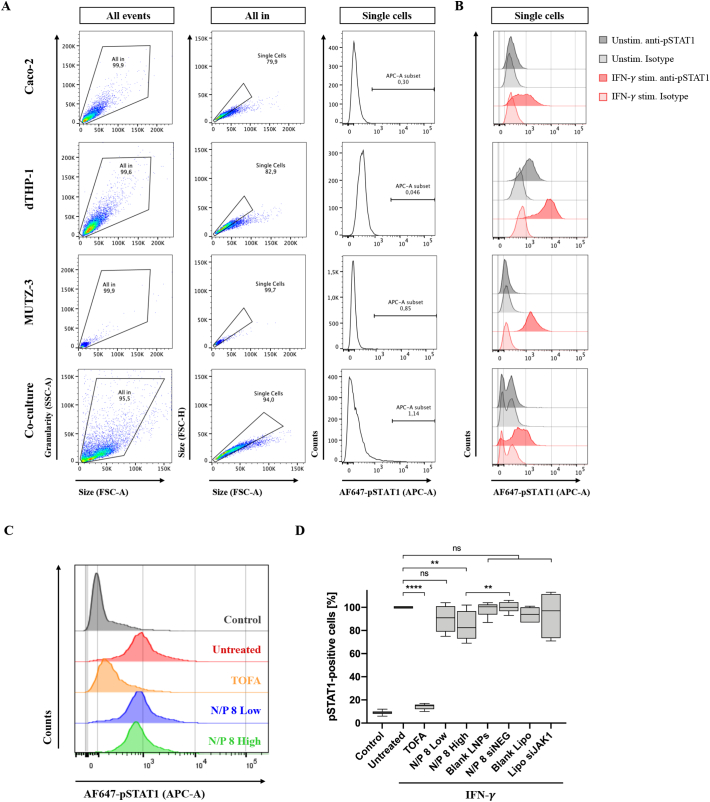
Fig. 6.
Flow cytometric analysis of JAK/STAT signal inhibition in leaky gut model. Mono-culture of Caco-2, dTHP-1, and MUTZ-3 cells or co-culture model were either left unstimulated or stimulated with IFN-γ (50 ng/mL) for 1 h. Cells were detached (mono-cultures) or dissociated (co-culture), fixed and permeabilized for intracellular staining with AF647 mouse anti-phospho-STAT1 (pY701) antibody or corresponding isotype control (AF647 mouse IgG 2a, κ). Flow plots show the gating strategy of unstimulated unstained controls for each cell line and co-culture model (A). Phosphorylation profile of STAT1 (pSTAT1) represented as histogram with percentage of pSTAT1-positive cells. The overlay of histograms summarizes (un-) stimulated controls stained either with specific pSTAT1 antibody (solid line) or isotype control (dashed line) (B). For JAK/STAT inhibition, the leaky co-culture model was pre-treated with TOFA (5 μM) or siJAK1-loaded nanoplexes (N/P ratio of 8) at low and high concentration (45 μg or 90 μg LNPs and 135 or 270 nM siRNA per well, respectively). Following the second 6 h sample incubation time, co-culture was IFN-γ stimulated, dissociated, and immediately stained and analyzed. Histogram overlay shows unstimulated untreated control (grey) and IFN-γ-induced pSTAT1 without anti-inflammatory treatment (untreated; red) or after treatment with TOFA (orange), or nanoplexes at low (blue) and high (green) concentration (C). Further control treatments included blank LNPs (90 μg/well); LNPs loaded with non-targeting siRNA (siNEG; 270 nM) and lipofectamine RNAiMAX (Lipo) alone or with siJAK1 at 270 nM concentration. pSTAT1-positive cells are represented as percentage, normalized to the untreated cells (D). The box plots depict the minimum and maximum values (whiskers), the upper and lower quartiles, and the median. Significant differences are represented as ** p < 0.01 and **** p < 0.0001 according to ANOVA; for TOFA (n = 15; N = 5), N/P 8 Low/High (n = 9; N = 3), and control treatments (n = 6; N = 2). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)