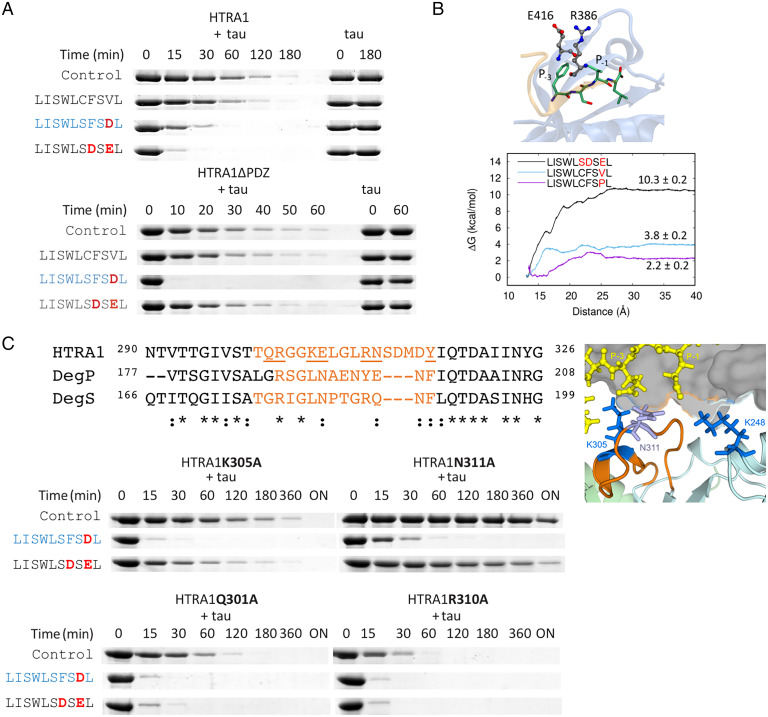
Fig. 4.
Effects of peptides derived from the C terminus of CAPN2 on HTRA1 activity. (A) Digests of soluble tau with HTRA1 and HTRA1ΔPDZ. 0.5 µM HTRA1 was incubated with 5 µM tau in 50 mM Tris pH 8.0 at 37 °C for the time points indicated in the absence (control) or presence of 50 µM of the peptides indicated. Deviations from the native CAPN2 sequence are indicated in red. The active-site ligand LISWLSFSDL is shown in blue. (B) Computational modeling and optimization of peptide sequences. Interaction between CAPN2 peptide and the PDZ domain. The mutation of residues P-1 and/or P-3 to Glu or Asp favors the formation of a salt bridge with Arg386 enhancing the binding of the peptide to the PDZ domain. The cartoon representation depicts the PDZ domain (blue) and peptide (beige), while the licorice representation shows the carbon atoms of residues of the PDZ domain (gray) and the peptide (green). Potential of mean force profiles, the reaction coordinate is the distance between the centers of mass of the PDZ domain and the peptides. (C) Loop L3 mutants implicated in the activation of HTRA1. Sequence alignment of the loop L3 sequences of HTRA1, DegP, and DegS. Activity of mutants determined by tau digests. 50 µM peptides were added when indicated. Cartoon model (Right) of LISWLSDSEL bound to the PDZ domain and its potential interactions with K305 and N311 of loop L3 and K248 of the protease domain based on high-resolution structures of the protease domain (PDB code: 3NZI) (loop L3, orange) and of the PDZ domain (gray) with the peptide DSRIWWV (yellow) (PDB code: 2JOA). Residues of loops of the protease domain pointing toward the bound peptide are shown as sticks.