Significance
In the dynamic environment of the airways, where SARS-CoV-2 infections are initiated by binding to human host receptor ACE2, mechanical stability of the viral attachment is a crucial fitness advantage. Using single-molecule force spectroscopy techniques, we mimic the effect of coughing and sneezing, thereby testing the force stability of SARS-CoV-2 RBD:ACE2 interaction under physiological conditions. Our results reveal a higher force stability of SARS-CoV-2 binding to ACE2 compared to SARS-CoV-1, causing a possible fitness advantage. Our assay is sensitive to blocking agents preventing RBD:ACE2 bond formation. It will thus provide a powerful approach to investigate the modes of action of neutralizing antibodies and other agents designed to block RBD binding to ACE2 that are currently developed as potential COVID-19 therapeutics.
Keywords: SARS-CoV-2, host–pathogen interactions, force spectroscopy, AFM, magnetic tweezers
Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections are initiated by attachment of the receptor-binding domain (RBD) on the viral Spike protein to angiotensin-converting enzyme-2 (ACE2) on human host cells. This critical first step occurs in dynamic environments, where external forces act on the binding partners and avidity effects play an important role, creating an urgent need for assays that can quantitate SARS-CoV-2 interactions with ACE2 under mechanical load. Here, we introduce a tethered ligand assay that comprises the RBD and the ACE2 ectodomain joined by a flexible peptide linker. Using magnetic tweezers and atomic force spectroscopy as highly complementary single-molecule force spectroscopy techniques, we investigate the RBD:ACE2 interaction over the whole physiologically relevant force range. We combine the experimental results with steered molecular dynamics simulations and observe and assign fully consistent unbinding and unfolding events across the three techniques, enabling us to establish ACE2 unfolding as a molecular fingerprint. Measuring at forces of 2 to 5 pN, we quantify the force dependence and kinetics of the RBD:ACE2 bond in equilibrium. We show that the SARS-CoV-2 RBD:ACE2 interaction has higher mechanical stability, larger binding free energy, and a lower dissociation rate compared to SARS-CoV-1, which helps to rationalize the different infection patterns of the two viruses. By studying how free ACE2 outcompetes tethered ACE2, we show that our assay is sensitive to prevention of bond formation by external binders. We expect our results to provide a way to investigate the roles of viral mutations and blocking agents for targeted pharmaceutical intervention.
A subset of coronaviruses (CoV) causes severe acute respiratory syndrome (SARS) in humans. We have seen three major recent outbreaks, including the first SARS pandemic from 2002 to 2004 (SARS-CoV-1), Middle East respiratory syndrome that emerged in 2012, and the ongoing COVID-19 pandemic (SARS-CoV-2). SARS-CoV-2 particles carry ∼100 copies of the trimeric viral glycoprotein Spike (S) on their surface (1), giving the appearance of an eponymous corona around the virus. Like SARS-CoV-1, SARS-CoV-2 attaches to human host cells by S binding to angiotensin-converting enzyme-2 (ACE2) (2–6) (Fig. 1A). Specifically, each of the three S1 subunits in an S trimer carries a receptor-binding domain (RBD) at its tip, which is presented in an up or down conformation and can bind ACE2 in the up conformation (Fig. 1B) (7). Binding of the virus to host cells occurs in dynamic environments (8, 9) where external forces act on the virus particle. In particular, in the respiratory tract, coughing, sneezing, and mucus clearance exert mechanical forces (10, 11) that the virus must withstand for productive infection. The magnitude and dynamics of these forces are not known precisely and are likely variable. A rough estimate from fluid dynamics suggests an upper limit of forces in the range of ∼2 pN to 2 nN (estimates are provided in SI Appendix).
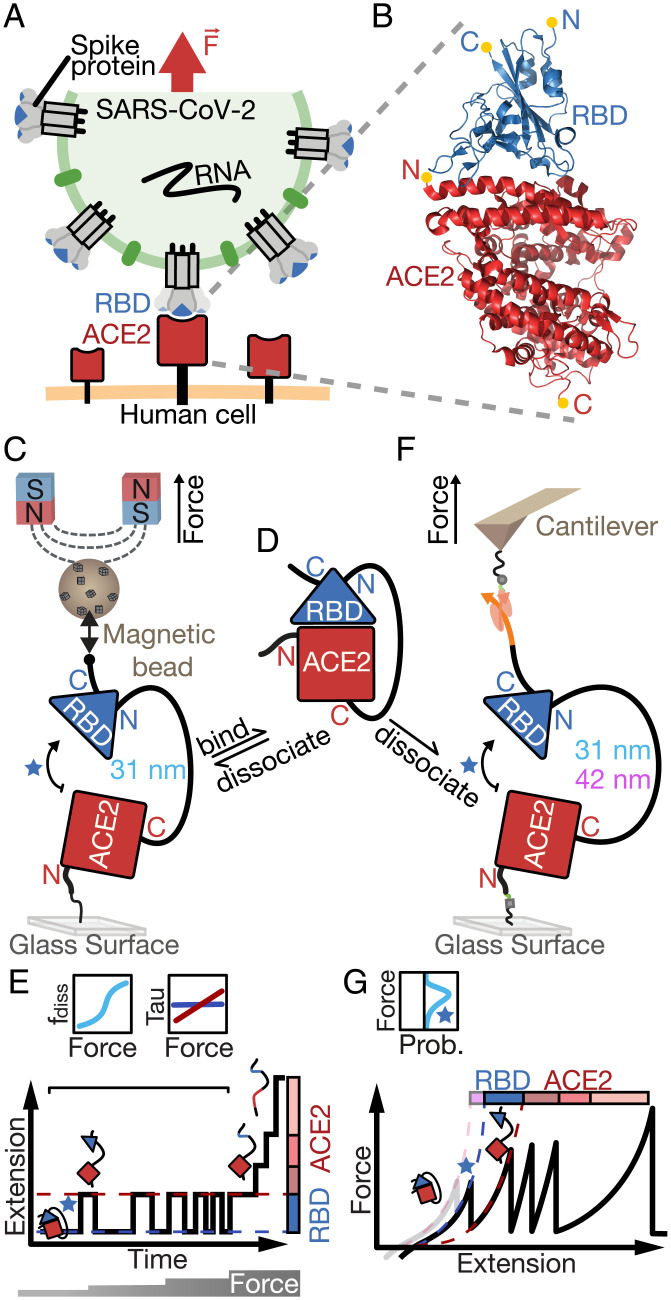
Fig. 1.
Single-molecule assays to probe the SARS-CoV-2 RBD:ACE2 interface under force. Motivation and overview of our tethered ligand assay for equilibrium and dynamic SMFS measurements in MT and AFM. (A) Schematic of a SARS-CoV-2 virus particle (green) presenting spike protein trimers (gray) that can bind to human ACE2 (red) on the cell surface via their RBDs (blue). The bond between RBD and ACE2 is formed in a dynamic environment, where it must withstand external mechanical forces (indicated by the red arrow), for example, caused by coughing or sneezing in the respiratory tract, in order to allow efficient infection of the human host cell (orange). (B) Crystal structure of the RBD:ACE2 complex (PDB ID: 6m0j). The N and C termini of the RBD (blue) and ACE2 (red) are indicated with yellow dots. (C) Schematic (not to scale) of the tethered ligand assay in MT. The tethered ligand construct consists of the ACE2 ectodomain (red square) and RBD (blue triangle) joined by a flexible polypeptide linker (black line) of 85 aa (31 nm contour length) or 115 aa (42 nm contour length). The tethered ligand construct is attached with one end covalently to the surface via an ELP linker (33) and with the other end to a superparamagnetic bead via a biotin–streptavidin bond. Permanent magnets above the flow cell enable the application of precisely calibrated stretching forces. (D) Tethered ligand construct in the absence of force, where the RBD remains bound to ACE2. (E) Stylized measurement of the tethered ligand construct in the MT. (Bottom) To probe RBD:ACE2 bond dynamics, time traces of the tether extension are recorded at different levels of applied force (indicated at the bottom). At low forces, reversible transitions between the bound configuration, with the RBD:ACE2 interface engaged, and a dissociated configuration, where the interface is broken and the peptide linker connecting the domains is stretched, are observed as jumps between two extension levels (red and blue dashed lines). At higher forces, further upward steps in the extension trace correspond to unfolding events of protein (sub)domains. (Top) From the MT time traces, both the fraction of time spent in the dissociated state and the dwell times in the bound and dissociated state can be determined as a function of applied force. (F) Schematic (not to scale) of the tethered ligand construct in the AFM. Here, covalent attachment to the surface uses a heterobifunctional PEG spacer, and the coupling to the AFM cantilever is accomplished via an Fgɣ tag on the protein that binds with very high force stability to the ClfA protein handle on the cantilever. (G) Stylized AFM measurement. (Bottom) The cantilever is retracted with constant velocity, and the force response to the applied extension is shown as a force-extension curve. With increasing extension, the RBD:ACE2 interface ruptures, protein subdomains unfold, and, finally, the ClfA:Fgɣ bond ruptures, giving rise to distinct peaks in the force-extension curve. Comparing two constructs with different linker lengths (31 nm black solid line and 42 nm gray/lilac alternative first peak) joining RBD and ACE2 allows assignment of the RBD:ACE2 interface rupture and unfolding of parts of the RBD to the first increment. Histograms of rupture forces (Top) are compiled from multiple measurements. The blue star refers to the RBD:ACE2 interface rupture.
The SARS-CoV-2 S protein and its interaction with ACE2 have been the target of intense research activity, as they are critical in the first steps of SARS-CoV-2 infection, and S constitutes a major drug and the key vaccine target in the current fight against COVID-19. Further, differences in binding between ACE2 and the SARS-CoV-1 and SARS-CoV-2 RBDs have been linked to the different observed patterns in lower and upper respiratory tract infections by the two viruses (5). Despite their importance, many questions about RBD:ACE2 interactions, particularly about their stability under external forces, are unresolved. Consequently, there is an urgent need for assays that can probe the affinity and kinetics of the interaction under a wide range of external forces. In nature, receptor–ligand pairs are often held in spatial proximity by neighboring interactions, creating high effective concentrations. Engagement of multiple interactions has been suggested to be important in other viral infections, including influenza, rabies, and HIV (12–16). Since conventional affinity measurements do not take into account these effects, there is a need for novel in vitro assays mimicking these effects when measuring bond characteristics.
Here, we present a tethered ligand assay to determine RBD interactions with ACE2 at the single-molecule level subject to defined levels of applied force. Our assay utilizes fusion protein constructs of SARS-CoV-1 or SARS-CoV-2 RBD and the human ACE2 ectodomain joined by flexible peptide linkers. To probe the linkage under a large range of mechanical forces and loading rates, we used two highly complementary single-molecule force spectroscopy (SMFS) approaches: an atomic force microscope (AFM) and magnetic tweezers (MT) (Fig. 1 C–G). We complemented the experiments with steered molecular dynamics (SMD) simulations to provide microscopic insights that are inaccessible experimentally.
AFM force spectroscopy can probe molecular interactions and protein stability dynamically (15–29), typically measuring at constant loading rate, and can investigate even the most stable high-force host–pathogen interactions (at forces F > 2,000 pN) (18). In AFM experiments, the molecular construct of interest is stretched between a surface and the tip of an AFM cantilever. The cantilever is retracted at a constant velocity, and the force is monitored from the cantilever deflection. Molecular rupture or protein (sub)domain unfolding events give rise to a sawtooth-like pattern in the force vs. extension traces (Fig. 1G). In contrast, MT typically operate at constant force and can resolve very low forces (19, 20), down to F < 0.01 pN. In MT, molecules are tethered between a flow cell surface and magnetic beads. External magnets apply defined and constant stretching forces, and the tether extension is monitored by video microscopy. In MT, unbinding or unfolding events give rise to steps in the extension vs. time trace (Fig. 1E).
Tethered ligand assays have provided insights into a range of critical molecular interactions under mechanical load (21–29). Under constant force, they allow observation of repeated interactions of the same binding partners, which are held in spatial proximity under mechanical control. Therefore, they can provide information on affinity, avidity, on and off rates, and mechanical stability (21, 23). Conversely, AFM force spectroscopy can perform dynamic measurements in a highly automated fashion and can reveal characteristic protein unfolding patterns, which can serve as molecular fingerprints (30) to select only properly folded and assembled molecular constructs for further analysis.
Probing our tethered ligand construct by AFM force spectroscopy, we reveal the dynamic force stability of the assembly. In combination with SMD simulations, we assign the increments revealed by force spectroscopy and establish the ACE2 unfolding pattern as a molecular fingerprint to select properly assembled tethers. Using MT, we measure the on and off rates at different levels of mechanical load and extrapolate to the thermodynamic stability at zero load. We compare the stability of the SARS-CoV-1 and SARS-CoV-2 RBD:ACE2 interactions in all three assays and consistently find a lower force stability for SARS-CoV-1 across the different techniques.
Results
A Tethered Ligand Assay to Probe Viral Attachment under Physiological Forces Using MT and AFM.
We designed tethered ligand fusion proteins that consist of the SARS-CoV-1 RBD or SARS-CoV-2 RBD and the ectodomain of human ACE2 joined by flexible polypeptide linkers (Fig. 1D). Protein constructs were designed based on the available crystal structures (31–33) of the SARS-CoV-1 or SARS-CoV-2 RBDs in complex with ACE2 and carry short peptide tags at their termini for attachment (Materials and Methods). For MT experiments, the protein constructs were coupled covalently to the flow cell surface via elastin-like polypeptide (ELP) linkers (33) and to magnetic beads via a biotin:streptavidin linkage (34). Tethering multiple proteins in MT enables parallel measurements of multiple molecules over extended periods (hours to weeks) at precisely controlled forces (34). In MT, bead positions and, therefore, tether extensions are tracked by video microscopy with ∼1 nm spatial resolution and up to kilohertz frame rates (35–37). For AFM experiments, we employ the same tethered ligand construct as used in the MT assay and covalently anchor it to a glass surface using polyethylene glycol (PEG) spacers. The key difference from the MT measurement is the use of an Fgɣ tag on the protein together with ClfA as a reversible handle system to attach it to the AFM cantilever. The ClfA:Fgɣ interaction is noncovalent, but can withstand extremely high forces of up to 2 nN, making it a reliable attachment modality with a built-in force fingerprint for AFM force spectroscopy (18). Together with stable custom-built AFM setups (38), this enables highly automated and reliable recordings of specific force-extension traces.
Dynamic AFM Force Spectroscopy Reveals a Characteristic Unfolding Pattern.
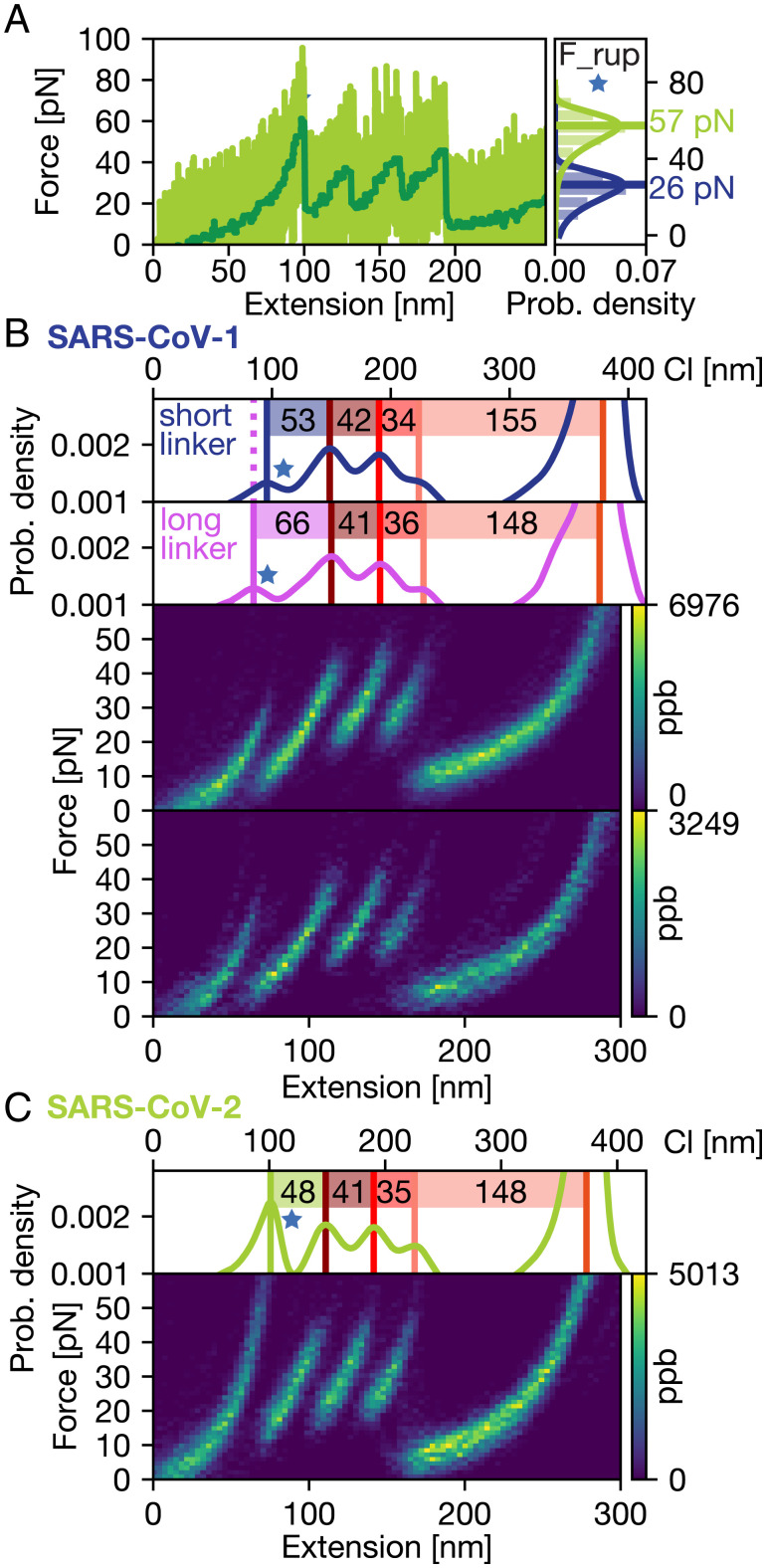
The AFM traces of the tethered ligand constructs feature a total of five sawtooth-like peaks, each corresponding to an unfolding or unbinding event (Figs. 1G and 2A and SI Appendix, Fig. S1). The last (rightmost) peak exhibits forces well within the range previously established for the ultrastrong ClfA:Fgɣ interaction (18), clearly indicating specific attachment of the protein construct. Consequently, the four peaks at lower forces must correspond to unfolding and unbinding events in a single tethered ligand construct. To visualize the most probable force-extension trace, we aligned and superimposed all individual curves (30, 39) (Fig. 2 B and C). They all feature the final rupture peak assigned to the ClfA:Fgɣ linkage and an initial (leftmost) peak typically at lower forces around 26 pN for the SARS-CoV-1 fusion construct and about twice as high forces for the SARS-CoV-2 fusion construct (around 57 pN; Fig. 2 A, Inset). This initial peak is followed by a trident-shaped, three-peak pattern at around 40 pN. The released contour lengths corresponding to each of these unbinding/unfolding peaks were determined from contour length transformations (30, 40) of each specific curve (see Materials and Methods). The four increments show the same order and very similar lengths for the SARS-CoV-1 and SARS-CoV-2 constructs (Fig. 2 B and C, Insets).
Fig. 2.
AFM force spectroscopy reveals multiple defined transitions in the tethered ligand construct. (A) AFM force-extension trace of the SARS-CoV-2 RBD:ACE2 tethered ligand construct (raw data in light green, total variation denoised in dark green, recorded at 800 nm/s retraction velocity). Four defined peaks are clearly visible between 30 and 60 pN. The final ClfA:Fgɣ rupture at >1,500 pN is not shown, for clarity. (B) Heatmaps of unfolding curves for the SARS-CoV-1 RBD:ACE2 fusion construct with short linker (31 nm, Top) and long linker (42 nm, Bottom) generated from 151/60 unfolding traces similar to the measurement shown in A. The color bar indicates the mapping of points per bin (ppb) values to each pixel in the heatmap. (C) Heatmap of unfolding curves for the SARS-CoV-2 RBD:ACE2 fusion construct generated from 127 unfolding curves, including the curve shown in A. Overall, the unfolding patterns look very similar for SARS-CoV-1 and 2; the only clear difference is that the first peak (indicated by a blue star throughout) is at higher forces for SARS-CoV-2. Inset at right in A directly compares the most probable rupture forces determined by the Bell–Evans model for the first peak for the two constructs. Insets above B and C show transformations of the force-extension data to contour length space (Materials and Methods). For SARS-CoV-1, two different length linkers were measured (31 nm linker in blue and 42 nm linker in purple); the difference of 13 nm in contour length for the initial peak is very close to the expected value.
The fact that the first peak (Fig. 2, blue star) has a notably different force signature for the SARS-CoV-1 and SARS-CoV-2 constructs, while all other peaks are very similar, suggests that the first peak involves the RBD domain, which is the only part that is different between the two constructs. To probe whether the first peak is due to the RBD:ACE2 interface opening or RBD unfolding, we performed control experiments with a longer linker (115 amino acids [aa] corresponding to 42 nm contour length, instead of 85 aa or 31 nm) between the two domains. The measurements with the longer linker reveal an increase of 13 nm in the contour length released in the first peak, very close to the expected 11 nm, which strongly suggests that the first peak represents dissociation of the RBD:ACE2 interface (Fig. 2B). The contour length increments of the first peaks (53 and 66 nm for the short and long linkers, respectively), are, however, larger than what is expected from interface opening only, which would release the linker lengths (31 nm or 42 nm) and involve reorientation of the domains. Conversely, full unfolding of the RBD domain would release 193 aa or 70 nm contour length, much longer than the increments that are experimentally observed. Therefore, the first peak must involve interface opening and partial unfolding of the RBD. Since all measurements were conducted under nonreducing conditions (Tris-buffered saline [TBS]; see Materials and Methods), we expect disulfide bridges between cysteines to be formed, which shield large parts of the RBD structure from force (31, 41) and allow only 51 aa to unfold, corresponding to 19 nm contour length, in excellent agreement with experimentally observed increments (SI Appendix, Fig. S2).
With the first peak assigned to the RBD:ACE2 interface opening and partial RBD unfolding, the subsequent trident-shaped, three-peak pattern is likely due to (stepwise) unfolding of the ACE2 domain. Control measurements with the ACE2 domain only and the same affinity tags resulted in traces showing the same trident-shaped pattern and no additional first peak (SI Appendix, Fig. S3), confirming the assignment of the trident-shaped pattern to the ACE2 domain. Comparing the same constructs with the SARS-CoV-1 and the SARS-CoV-2 RBD tethered to ACE2 reveals a lower force stability for the SARS-CoV-1 interface compared to SARS-CoV-2 (Fig. 2A, Insets). In addition, the AFM data suggest that, after the opening of the interface, stepwise ACE2 unfolding gives rise to a defined pattern that can be used as a molecular fingerprint.
All-Atom SMD Simulations Provide Insights into the Unfolding Patterns in Molecular Detail.
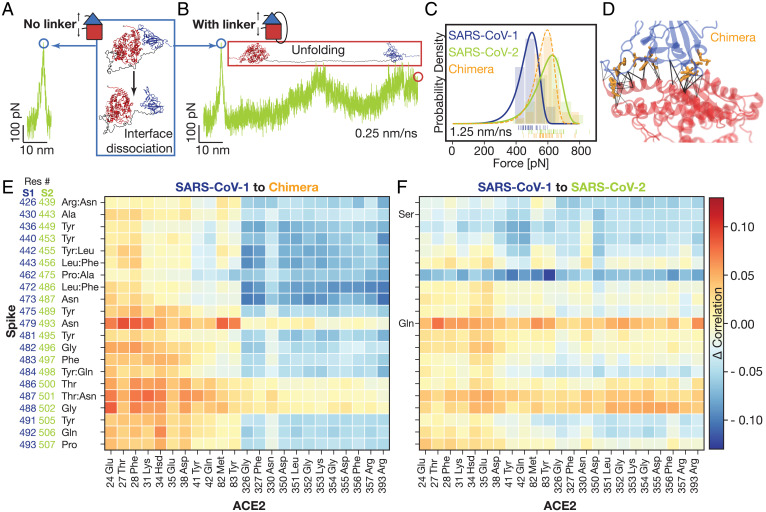
In an in silico SMFS approach, the tethered ligand protein probed in the AFM measurement was modeled using QwikMD (42). Based on the available crystal structures, the RBD:ACE2 complex was modeled with and without the polypeptide linkers for both SARS-CoV-1 and SARS-CoV-2, in a total of four different systems (for details, see Materials and Methods). Over 300 SMD simulations were performed employing GPU-accelerated NAMD 3 (43). The simulated behavior of the complexes agrees with the AFM experiments, starting with the dissociation of the RBD domain from ACE2 and consecutive unfolding of the RBD (Fig. 3 A and B). In the simulations, the unfolding of parts of the RBD is caused by the linker that gets stretched after the interface is released from ACE2. After the initial RBD dissociation, ACE2 unfolds in several substeps. The corresponding force–distance curve (Fig. 3B) agrees with the observed unfolding pattern obtained by AFM measurements. Fig. 3 A and B, Inset shows renderings at three different time points to visualize the forced RBD:ACE2 dissociation observed in SMD and AFM experiments.
Fig. 3.
SMD simulations. (A) SMD force-extension curve for pulling on SARS-CoV-2 RBD:ACE2 interaction without additional linker. Inset (blue frame) shows a rendering of the interface opening causing the peak in the trajectory. (B) SMD force-extension curve for pulling on tethered ligand SARS-CoV-2 RBD:ACE2 construct (same complex as investigated with the AFM and MT). Force and pathway of interface opening (indicated with blue circle) are equivalent to pulling on the interaction without linker as shown in A. Inset (red frame) shows rendering at the end of the simulation, when interface is dissociated and ACE2 and RBD are partially unfolded. (C) Force distribution of interface opening for SARS-CoV-1 (most probable rupture force: 499 pN), SARS-CoV-2 (most probable rupture force: 628 pN), and chimera (most probable rupture force: 597 pN) tethered ligand constructs. Chimera is a tethered ligand construct with the SARS-CoV-1 RBD with 12 aa replacements according to the SARS-CoV-2 RBD. These 12 aa replacements in the SARS-CoV-1 RBD nearly reproduce force stability of SARS-CoV-2. Distributions are from 40 replicas for each construct. (D) Crystal structure of the SARS-CoV-1 RBD:ACE2 interaction (PDB-ID: 2ajf). Residues changed for chimera are indicated in orange. Black lines quantify correlation between residues from the RBD and ACE2 shortly before interface rupture for the chimera system. The overall correlations between RBD and ACE2 residues determine the force propagation pathways through the interface and thus the stability of the interaction. (E and F) Heatmaps of the change in correlation of individual spike residues with ACE2 residues comparing SARS-CoV-1 with the chimera (E) and SARS-CoV-1 with SARS-CoV-2 (F). Residues were selected based on proximity to the interface. Modified residues in the chimera in close proximity with ACE2 are indicated in the y axis labels. Heatmaps indicate a change in mechanostability pattern throughout the interface. Difference in the interaction pattern under force load between SARS-CoV-1 and SARS-CoV-2 RBD:ACE2 interface helps to rationalize their difference in stability.
Simulations of the RBD and ACE2 without and with the linker showed identical behavior until the point of the interface dissociation (Fig. 3 A and B), and the dissociation process was conserved for both protein models. The rupture forces of SARS-CoV-1 and SARS-CoV-2 were determined both for the tethered ligand protein and for just the single domains. For collecting statistics, the construct without linker was used to determine unbinding forces of the RBD:ACE2 interface. The simulations give 26% higher forces for SARS-CoV-2 compared to SARS-CoV-1 (Fig. 3C). The observed higher forces for SARS-CoV-2 qualitatively agree with the differences in forces determined from the AFM measurements, with an expected increase in absolute forces due to the much higher loading rates in the SMD simulation compared to AFM measurements.
Unfolding Patterns across Different Force-Loading Rate Regimes Are Highly Reproducible.
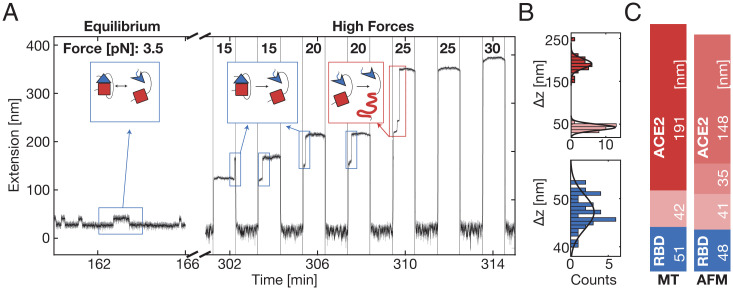
In contrast to AFM and SMD simulations, where tethered fusion constructs were subjected to forces with a constant force-loading rate, MT were used to examine the fusion complex at constant forces, first studying equilibrium binding and dissociation between RBD and ACE2 at lower forces and then unfolding individual protein domains at high forces (Fig. 4A). At forces between 2 pN and 5 pN, we observe systematic transitions in the extension traces, with jumps between a high extension “dissociated” and low extension “bound” state (Fig. 4A, “Equilibrium” and Fig. 5 A and B). The transitions systematically shift toward the dissociated state with increasing force (Fig. 5A). Alternating between levels of low force (0.5 pN) and higher forces (15, 20, 25, and 30 pN; Fig. 4A, “High Forces”) reveals three distinct unfolding transitions. At forces of 15 and 20 pN, one unfolding transition repeatedly occurs after refolding during a low-force interval. This reversible transition corresponds to the systematic transitions recorded at equilibrium between 2 and 5 pN (Fig. 4A, blue boxes and Fig. 4B, blue histogram for high-force transitions), while a subsequent two-step transition above 25 pN (Fig. 4A, red box and Fig. 4B, red histograms) is irreversible. Measuring just the ectodomain of ACE2 revealed the same characteristic irreversible two-step unfolding pattern above 25 pN without showing the systematic transitions at lower forces or the reversible unfolding at 15 and 20 pN (SI Appendix, Fig. S4). This suggests a clear assignment of the two irreversible unfolding events to the unfolding of ACE2, while the equilibrium transitions, observed as reversible steps at 15 and 20 pN, are attributed to the RBD:ACE2 interface opening and partial RBD unfolding, corresponding to the first peak in the AFM force–distance curves.
Fig. 4.
SARS-CoV-2 RBD:ACE2 interface opening and unfolding in MT. (A) Extension-time trace of an RBD:ACE2 tethered ligand construct in MT shows distinct transitions at different levels of constant force. At low forces (labeled “Equilibrium”), we observe stochastic transitions between two extension levels, separated by ΔRBD ≈ 13.5 nm at a force of 3.5 pN. In subsequent measurements at higher forces (labeled “High Forces”), the force is iteratively altered between 0.5 pN (low extension) and increasingly higher forces (indicated at the top). At 15 and 20 pN, the RBD:ACE2 interface ruptures together with a partial RBD unfolding (ΔRBD ≈ 32 nm) and refolds in the subsequent 0.5 pN interval. At 25 pN, ACE2 irreversibly unfolds in two steps, first a smaller one (ΔACE2small ≈ 30 nm) and then a larger one (ΔACE2large ≈ 135 nm). Gray trace, five-frame moving average filtered; black trace, 40 frame moving average filtered. (B) Histograms of unfolding increments at high forces of the RBD (blue) and the two parts of ACE2 (pink, red) in MT. The histograms are fitted with Gaussians (solid black lines). Some points are outside of the plotting range for clarity but included in the fit. (C) Mean unfolding increments observed for tethered ligand construct with constant forces in MT and with constant loading rate in AFM. The observed increments were transformed into contour lengths using the WLC model, assuming a persistence length of 0.5 nm in MT and 0.365 nm in the AFM. Values ± SD for the increments in MT are ΔRBD = (51 ± 10.8) nm, ΔACE2_1 = (42 ± 5.5) nm, and Δ ACE2_combined = (191 ± 18.7) nm. Values for the increments in the AFM are ΔRBD = 48 nm, ΔACE2_1 = 41 nm, Δ ACE2_2 = 35 nm, and ΔACE2_3 = 148 nm. Values in B and C correspond to mean values from 42 molecules observed in MT and 127 molecules observed in AFM.
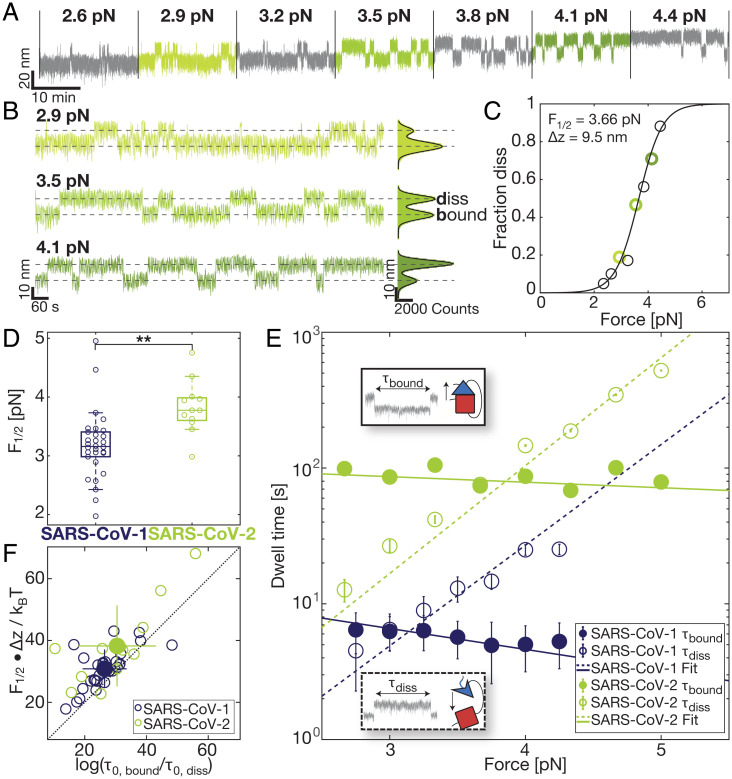
Fig. 5.
Comparison of mechanical stability and kinetics of ACE2 binding to SARS-CoV-1 and SARS-CoV-2 RBD. (A) Extension-time traces at different constant forces for the SARS-CoV-2 RBD:ACE2 fusion construct reveal stochastic transitions between two extension levels, corresponding to the bound and dissociated RBD:ACE2 interface. Increasing the force increases the fraction of time spent in the dissociated conformation. (B) Expanded views of extensions at three different forces: below, at, and above the midpoint force from the trace in A show shifts toward the dissociated conformation with increasing force; same color code as in A. The dissociated and bound states are indicated by dashed lines. The histograms of the extensions are fitted with a double Gaussian (solid black lines). (C) The fraction of time in the dissociated conformation determined from extension-time traces (symbols; points determined from the traces in B are shown with matching color codes). The black line is a fit of Eq. 1. (Inset) Fitting parameters F1/2 and Δx. (D) Comparison of the F1/2 distribution between SARS-CoV-1 and SARS-CoV-2 reveals a significantly higher force stability of the SARS-CoV-2 RBD:ACE2 bond (P = 0.00124; two-tailed t test; significance indicated by two stars). Data points are the fitted F1/2 from independent molecules. Boxes are the median and interquartile range. (E) Dwell times in the dissociated (open symbols) and bound state (filled symbols) determined from extension-time traces for SARS-CoV-1 (blue) and SARS-CoV-2 (green). Mean dwell times for individual molecules were determined from maximum likelihood fits of a single exponential to the dwell time distributions. The symbols and error bars are the mean and SD from log-averaging over 29 (SARS-CoV-1) and 12 (SARS-CoV-2) molecules. Dashed and solid lines correspond to the mean of the exponential fits to the individual dwell times in the bound state and dissociated state, respectively. Insets visualize dwell times in MT extension-time traces. (F) Free energy differences between the bound and dissociated state of the RBD:ACE2 tethered ligand constructs. The free energy differences were obtained from the equilibrium data as F1/2⋅Δx and from the dynamics as log(τ0,bound/τ0,diss). The data fall along the 45° line (dashed), indicating that the two estimates give consistent values. Comparison of the SARS-CoV-1 and SARS-CoV-2 data reveals a larger free energy difference for SARS-CoV-2, indicating a more stable interface. Distributions and mean values shown in D–F are for 29 (SARS-CoV-1) and 12 (SARS-CoV-2) molecules, respectively.
Unlike in the SMD simulations, where interface dissociation clearly precedes partial RBD unfolding, the processes can be discriminated within the resolution limits neither of the AFM nor of MT. Thus, we are unable to clearly assign a temporal order or causality in the in vitro experiments. If the process in the AFM and MT was equal to what SMD simulations showed, the linker would be unstretched upon interface dissociation. It is highly likely that the subsequent partial RBD unfolding should be observable as a separate event at the loading rates in the in vitro experiments. The fact that both processes are not separable in both techniques suggests that the clear order suggested by SMD simulations is an artifact of the high force-loading rates and that, in the experiments, partial RBD unfolding might happen simultaneously or even precede interface dissociation.
Using the worm-like chain (WLC) model (44, 45) with a persistence length of 0.5 nm (34, 46), the contour lengths of the observed extension increments in MT are calculated (Fig. 4B). The mean values of the increments in MT (Fig. 4C, “MT”) are compared to the contour length transformed increments in the AFM (Fig. 4C, “AFM”; all values are given in SI Appendix, Table S1). Strikingly, the increments observed with both SMFS techniques are in excellent agreement, within experimental errors, with each other and with theoretical expectations based on the crystal structure (31) and the fusion construct design. Comparing the total length gain from unfolding the tethered ligand ACE2 in the AFM (224 nm) and MT (233 ± 25.5 nm) reveals quantitative agreement within experimental errors. The first ACE2 unfolding event (SI Appendix, Table S1, ACE2_1) in the AFM almost perfectly matches the first increment in MT. The second and third ACE2 unfolding (SI Appendix, Table S1, ACE2_2 and ACE2_3) in the AFM cannot be separately resolved in MT for most molecules and typically occur as one single large step (SI Appendix, Table S1, ACE2_combined). In a small subpopulation (8 of 42 total molecules), however, we observed a very short-lived intermediate level, splitting the large step into a smaller and yet another larger step, matching the increments observed in the AFM. In summary, ACE2 unfolding provides a highly specific and reproducible molecular fingerprint across AFM and MT measurements that we subsequently used to select for specific tethers to probe the RBD:ACE2 interface.
MT Measurements Probe the RBD:ACE2 Interaction under Load in Equilibrium.
After selecting specific tethers based on the ACE2 fingerprint, we analyze the equilibrium transitions measured in MT at forces between 2 and 5 pN. The transitions systematically change with applied force: At low forces, the interface is predominantly formed (bound state), while increasing the force increases the fraction of time with an open interface and a partially unfolded RBD (dissociated state) (Fig. 5A). Histograms of the tether extension revealed two clearly separated peaks that are fit well by a double Gaussian (Fig. 5B, solid black lines). By setting thresholds at the minimum between the extension peaks, we defined populations in the dissociated and bound states. The fraction in the dissociated state (Fig. 5C, circles) follows a sigmoidal force dependence. The data are well described by a two-state model (Fig. 5C, solid line) where the free energy difference between the two states (bound vs. dissociated but still attached to each other with a linker) depends linearly on the applied force F, that is, ΔG = ΔG0 –F⋅Δz, such that the fraction in the dissociated state is
| [1] |
where kB is Boltzmann’s constant and T is the absolute temperature. F1/2 and Δz are fitting parameters that represent the midpoint force, where the system spends half of the time in the dissociated and half of the time in the bound conformation, and the distance between the two states along the pulling direction, respectively. The free energy difference between the bound and dissociated states at zero force is given by ΔG0 = F1/2 ⋅ Δz and provides a direct measure of the stability of the binding interface.
From fits of Eq. 1 to the data for the SARS-CoV-2 RBD:ACE2 construct, we found F1/2 = 3.8 ± 0.4 pN and Δz = 10.2 ± 3.7 nm, and, therefore, ΔG0 = F1/2 ⋅ Δz = 5.5 ± 2.1 kcal/mol (data are the mean and SD from fits to biological repeats; see SI Appendix, Table S2 for a summary of all fitted parameters). The value of Δz determined from fitting Eq. 1 is within experimental error in agreement with the distance between the dissociated and bound states ΔzG = 13.5 ± 1.8 nm determined from fitting two Gaussians to the extension histograms at the equilibrium force F1/2 and evaluating the distance between the fitted center positions. The observed Δz is also in excellent agreement with the expected extension change of ∼13.4 nm, based on the crystal structure (31) (Protein Data Bank [PDB] ID: 6m0j) taking into account the stretching elasticity of the 85 aa protein linker and the unfolding of nonshielded parts of the RBD using the WLC model (34, 45, 46) with a bending persistence length of Lp = 0.5 nm and assuming 0.365 nm/aa.
In addition to providing information on the binding equilibrium, the MT assay gives access to the binding kinetics under force. Analyzing the extension-time traces using the same threshold that was used to determine the fraction dissociated vs. force, we identify dwell times in the dissociated and bound states (SI Appendix, Fig. S5 A and B), which are exponentially distributed (SI Appendix, Fig. S5 C and D). The mean dwell times in the dissociated state increase with increasing force, corresponding to the intuitive interpretation that the higher the force, the longer it takes for a dissociated receptor:ligand pair to rebind (Fig. 5E, dashed green line). The mean dwell times in the bound state, on the other hand, decrease with increasing force, albeit only slightly, implying that the higher the force, the shorter the bond stays intact. The dependencies of the mean dwell times on the applied force F are well described by exponential, Arrhenius-like relationships (47),
| [2] |
where the fitting parameters τ0,diss and τ0,bound are the lifetimes of the dissociated and bound conformation in the absence of force, and Δzdiss and Δzbound are the distances to the transition state along the pulling direction.
The parameters Δzdiss and Δzbound quantify the force dependencies of the lifetimes of the respective states, and the slopes in the log(τdiss/bound) vs. F plots (Fig. 5E) are given by Δzdiss/bound/kBT. Δzbound is smaller than Δzdiss (by more than a factor of ∼5); that is, dissociation of the bound complex is less force sensitive than rebinding from the dissociated conformation. The different force sensitivities can be rationalized from the underlying molecular processes: The bound complexes feature protein–protein interactions that will break over relatively short distances; in contrast, the dissociated conformations involve flexible peptide linkers that make rebinding from the dissociated states more force dependent. The sum of Δzdiss and Δzbound is 7.9 ± 5.1 nm and, within experimental error, equal to Δz obtained from fitting Eq. 1, which indicates a single major energy barrier separating the bound and the dissociated states.
The extrapolated lifetimes at zero force of the bound conformations τ0,bound are in the range of 115 s for SARS-CoV-2. In comparison, the lifetimes of the dissociated states in the absence of load τ0,diss are much shorter, similar to ∼0.07 s (SI Appendix, Table S2). The extrapolated lifetimes at zero force provide an alternative route to computing the free energy difference between the dissociated and bound states at F = 0, which is given by ΔG0,tau = kBT ⋅ log(τ0,diss/τ0,bound). We find good agreement, within experimental error, between the free energy differences ΔG0,tau determined from the extrapolated lifetimes and the values ΔG0 = F1/2 ⋅ Δz from Eq. 1 (SI Appendix, Table S2 and Fig. 5F), providing a consistency check between equilibrium and kinetic analysis. The results show that our tethered ligand assay in MT can yield consistent information on the binding equilibrium and on the interaction kinetics under external force.
Comparing the extrapolated lifetimes at zero force and the distances to the transition state obtained from MT measurements to values obtained from fitting a dynamic force spectrum recorded with the AFM yields reasonable agreement (SI Appendix, Fig. S6). This suggests that both techniques probe similar pathways.
SARS-CoV-2 Attachment Is More Stable and Longer Lived than SARS-CoV-1 under Constant Load.
A construct using the same 85 aa linker and attachment geometry, but the SARS-CoV-1 RBD instead of SARS-CoV-2 RBD, shows a qualitatively very similar force response in MT, with stochastic transitions between a bound and a dissociated conformation at an equilibrium force below 5 pN and ACE2 unfolding at forces higher than 25 pN. The increments from unfolding the tethered ACE2 are in excellent agreement with the increments both from the ACE2 unfolding of the SARS-CoV-2 tethered ligand construct in MT and from the AFM ACE2 unfolding (SI Appendix, Fig. S4). Furthermore, they happened at comparable forces to those in the SARS-CoV-2 tethered ligand construct and single ACE2 in MT. As for the SARS-CoV-2 tethered ligand construct, the molecules for equilibrium and kinetic analysis of the SARS-CoV-1 RBD:ACE2 bond could thus be selected based on the molecular fingerprint of ACE2 unfolding and directly compared to SARS-CoV-2 tethered ligand constructs.
From fits of Eq. 1, we found F1/2 = 3.2 ± 0.6 pN, Δz = 9.7 ± 1.7 nm, and thus ΔG0 = 4.4 ± 1.1 kcal/mol for the SARS-COV-1 RBD:ACE2 tethered ligand construct (SI Appendix, Table S2). The length increment Δz is, again, within experimental error, in good agreement with the value determined from fitting two Gaussians to the extension histogram near the midpoint of the transition (ΔzG = 11.3 ± 1.7 nm at F1/2). The slightly shorter extension increment upon dissociation for the SARS-CoV-1 construct compared to SARS-CoV-2, despite using the same 85 aa linker and a very similar crystallographic geometry, is mostly due to the smaller extension of the WLC at the lower midpoint force for SARS-CoV-1. Comparing the two lengths after contour length transformation yields ΔzSARS-CoV-1 RBD, WLC = 51.2 ± 7.9 nm and ΔzSARS-CoV-2 RBD, WLC = 50.3 ± 7.7 nm, in agreement, within error. In contrast, the SARS-CoV-1 RBD:ACE2 interface shows a significantly lower midpoint force F1/2 than the SARS-CoV-2 RBD:ACE2 bond (Fig. 5D; P = 0.00124 from a two-tailed t test), in line with the lower unbinding forces for SARS-CoV-1 observed in the AFM (Fig. 2 B and C) and SMD simulations. The difference also manifests in a reduced mean free energy of the SARS-CoV-1 RBD:ACE2 bond computed from both the midpoint forces and distances as well as from the rates (Fig. 5F). Comparing the lifetimes of the bound and dissociated states (Fig. 5E, blue and green lines) and extrapolating the mean lifetimes to zero force yields no significant difference between the lifetimes in the dissociated conformation (Fig. 5E, dashed lines, τ0,diss, SARS-CoV-1 = 0.03 ± 0.05 s, τ0,diss, SARS-CoV-2 = 0.07 ± 0.19 s, P = 0.293). The extrapolated lifetime in the bound state at zero force, on the other hand, is significantly different at α = 0.1 (τbound, SARS-CoV-1 = 19.0 ± 24.8 s, τbound, SARS-CoV-2 = 114.5 ± 278.8 s, P = 0.072) and more than 6 times higher for SARS-CoV-2, indicating a lower dissociation rate from ACE2. Furthermore, the lifetime of the SARS-CoV-2 RBD:ACE2 bond decreases less with force, that is, has a shallower slope than the bond lifetime of SARS-CoV-1 RBD:ACE2. Therefore, the longer lifetime of the SARS-CoV-2 bond compared to SARS-CoV-1 becomes even more pronounced under mechanical load.
MT Provide a Sensitive Assay to Study Molecules that Block the RBD:ACE2 Interaction.
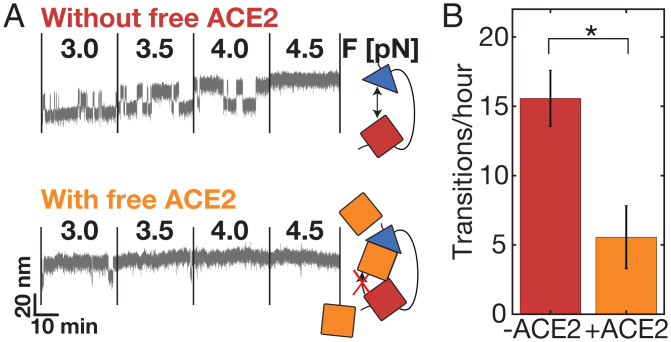
Apart from providing a tool to assess equilibrium binding and kinetics, investigation of the tethered ligand assay in MT also allows us to probe the influence of other binding partners on the bond dynamics. For this purpose, the equilibrium bond dynamics are first recorded under standard conditions before exchanging the buffer and conducting the same experiment in the presence of the compound of interest. As a proof of concept, we investigate the influence of soluble ACE2 on the bond dynamics (Fig. 6). First, we recorded the equilibrium binding of the SARS-COV-2 RBD and the tethered ACE2 in a measurement between 3.0 and 4.5 pN, observing the characteristic transitions between the dissociated and bound conformations (Fig. 6A, “Without free ACE2”). We then added 3.8 µM free, soluble ACE2 and increased the force to 7 pN, to ensure the dissociation between the tethered receptor–ligand pair and enable binding of free ACE2, before conducting the same measurement in the presence of the free ACE2 (Fig. 6A, “With free ACE2”). At the same forces, the system is now predominantly in the dissociated conformation, rebinding only occasionally. This matches the interpretation of free ACE2 binding to the RBD and thus preventing the tethered ACE2 from binding and transitioning into the bound conformation. Overall, we find that the number of dissociation and rebinding events is significantly reduced in the presence of free ACE2 (Fig. 6B; P = 0.022 from a repeated measures t test over six independent molecules).
Fig. 6.
Blocking of the ACE2:SARS-CoV-RBD bond with free ACE2. (A) Extension–time traces of a SARS-CoV-2 tethered ligand construct in MT around the equilibrium force, first without and then in the presence of free ACE2 (∼3.8 µM) in solution. (Top) Without soluble ACE2, the RBD:ACE2 bond frequently dissociates and rebinds (corresponding to changes in extension level). (Bottom) In the presence of free ACE2, the bond is significantly longer dissociated already at low forces, rebinding only a few times (corresponding to a longer time spent at the level of high extension). (B) Quantification of mean number of transitions per hour during the force ramp shown in A with and without free ACE2 for six independent molecules. Error bars indicate SEM. The difference is statistically significant with P = 0.0218 (two-tailed t test for two dependent means).
Molecular Modeling Provides Insights into the Molecular Origin of Higher Mechanostability of SARS-CoV-2.
In all simulations and experiments, the SARS-CoV-2 RBD:ACE2 complex was shown to be more resilient to mechanical load than the same complex from SARS-CoV-1. To investigate the origin of the higher mechanostability of the SARS-CoV-2 complex, we designed an in silico protein chimera by combining the SARS-CoV-1 RBD core with SARS-CoV-2 RBD residues located on the RBD:ACE2 interface. Our protein chimera was built using a homology modeling approach, similar to the protocol previously developed to investigate mechanostable protein folds (48). We compared the sequence and structural differences between SARS-CoV-1 and SARS-CoV-2 by investigating both RBD:ACE2 interfaces in PDBsum (49) and VMD (50). While both RBDs have high structural similarity (with an rmsd of 1.39 Å), they only share 72% sequence identity, revealing a total of 64 residue changes between SARS-CoV-1 and SARS-CoV-2 RBDs. After selecting the RBD amino acid residues that form the interface with ACE2 in both structures, we identified 12 residues that were different in the two SARS’ RBDs. Using the structure of SARS-CoV-1 as a template (PDB ID: 2ajf), Modeler (51) was employed to mutate the 12 selected residues (SI Appendix, Table S3), to create a protein chimera that is identical to the SARS-CoV-1 sequence except for the 12 mutations that make the interface sequence similar to that of SARS-CoV-2 (Fig. 3D).
The RBD chimera enables us to investigate the impact of the interface residues on the mechanism of force stability of the RBD–ACE2 complex (Fig. 3 C–F). Although interface residues are key players in the binding affinity of proteins (52), the same does not necessarily hold true for protein interface mechanostability, where unbinding pathways are frequently determined by complex force propagation profiles (53, 55). Strikingly, SMD simulations of the chimeric protein reveal rupture forces that are only 5% weaker than the SARS-CoV-2 complex (Fig. 3C), suggesting that the 12 amino acid substitutions in the RBD of SARS-CoV-1 are enough to achieve force resilience similar to the SARS-CoV-2 complex. This observation is noteworthy because it shows that the modified surface residues alone were enough to significantly change the complex's resistance to mechanical stress.
To probe the contribution of the individual amino acid modifications, we employed a generalized correlation approach (54, 56) to produce a detailed analysis of the contact network during the pulling simulations. In short, we calculate the correlation between the movement of residues to determine how cooperative their motion is. The higher the correlation, the more relevant their interaction is for the stability of the complex (Fig. 3 E and F). This analysis revealed that, at high forces, only 7 of these 12 amino acids remain in close proximity to ACE2, directly contributing to the stabilization of the complex. The seven modifications are indicated in Fig. 3E. Moreover, the analysis revealed that the 12 substitutions drastically increase the overall correlation between the spike residues and the two alpha helices of ACE2’s contact surface (SI Appendix, Fig. S7). Furthermore, the analysis also revealed that residue N479 in the chimera and its equivalent Q493 in the SARS-CoV-2 RBD plays a much bigger role in stabilizing the complex than in the SARS-CoV-1 RBD. This amino acid residue has been shown to be a key player in the binding of RBD to ACE2 (57). We could also observe that correlations between residues from the chimera and ACE2 consistently increased in the region around modified residue N487. The equivalent residue in SARS-CoV-2 RBD, N501, is mutated in several SARS-CoV-2 variants of concern. We note that, for SARS-CoV-2, the interactions between RBD and the N-terminal helices of ACE2 (residues 24 to 83) are found to be significantly more stable than for SARS-CoV-1, as indicated by the stronger correlations in this region (Fig. 3 E and F).
Discussion
In order to infect human host cells, SARS coronaviruses engage with receptors in the turbulent and dynamic environment of the respiratory tract, where forces impact their anchoring. Force stability of the RBD:ACE2 bond thus plays a crucial role for the virus to be able to infect host cells efficiently. Understanding the binding mechanism and being able to assess the stability of the RBD:ACE2 bond—also under the influence of external agents—opens opportunities to develop and test drugs and predict fitness advantages of mutated RBDs. Here, we have reported a tethered ligand assay to study and characterize the stability and binding mechanism of the SARS-CoV RBD:ACE2 bond with single-molecule precision.
Using the unfolding of ACE2 as a fingerprint pattern, we were able to bring together equilibrium studies using MT, high-force rupture analysis carried out with an AFM, and molecular insights of the unfolding process obtained by SMD simulations. Combining these approaches, we were able to cover a large range of physiologically relevant forces. We compared the force stability of the bond of the SARS-CoV-1 and SARS-CoV-2 RBD to human receptor ACE2 and found higher force stability for the novel coronavirus throughout all force regimes. This is in line with previously published in silico and in vitro force spectroscopy studies that found a 20 to 40% difference in the force stability of SARS-CoV-1 and SARS-CoV-2 RBD:ACE2 interactions (56, 57). The higher force stability of SARS-CoV-2 engaging ACE2 might contribute to the fact that SARS-CoV-2 more frequently infects the upper respiratory tract in addition to deep lung tissue compared to the 2002 SARS variant (8, 10), which, in turn, appears to increase its population spread and to make the new virus harder to contain. Higher force stability might, therefore, increase transmissibility, which could also play a role in understanding fitness advantages of newly evolving SARS-CoV-2 variants.
In addition to force stability, we analyzed the binding kinetics of the complex under force obtained by MT measurements, finding an order of magnitude higher bond lifetime of the SARS-COV-2 RBD construct compared to the same construct with the SARS-COV-1 RBD at their respective equilibrium forces. Extrapolating these lifetimes to zero force yields a more than fivefold higher bond lifetime (∼115 s) for SARS-CoV-2 than for SARS-CoV-1 (∼20 s). We can quantitatively relate our results to studies that have reported equilibrium dissociation constants and rates for the ACE2 interactions with SARS-CoV-1 and SARS-CoV-2 using traditional binding assays. While the values reported in the literature vary significantly, likely due to the different experimental methods and sample immobilization strategies used, clear and consistent trends can be identified. The zero-force lifetimes of the bound complex determined in our assay correspond to rates of k0,off ∼5 ⋅ 10−2 s−1 for SARS-CoV-1 and ∼8.7 ⋅ 10−3 s−1 for SARS-CoV-2, well within the ranges of reported ksol,off values in literature (6, 31, 58–61) (for an overview, see SI Appendix, Table S4). Our value for the off rate of SARS-CoV-2 RBD bound to ACE2 is also in excellent agreement with the value of (8 ± 5) ⋅ 10−3 s−1 extrapolated from previous AFM force spectroscopy experiments (62). In contrast to the difference in off rates between SARS-CoV-1 and SARS-CoV-2, the (bimolecular) on rates reported in literature are similar for both SARS variants, in the range of ∼105 M−1⋅s−1. Consistently, our tethered ligand assay finds similar unimolecular on rates on the order of ∼10 s−1. Even though both bulk and single-molecule assays find similar on rates for the two SARS viruses, the absolute values are not comparable, as solution-based assays determine bulk on rates (with units of molars per second), whereas our tethered ligand assay measures molecular on rates (in per second). To relate the two quantities, an effective concentration ceff can be introduced, such that ksol,on = k0,on/ceff (21, 63, 64). The data imply an effective concentration at zero force on the order of 35 µM for the construct with an 85 aa linker, in the range of concentrations found for other tethered ligand protein systems (64, 65). Under force, the complex will spend more time in the dissociated state (i.e., the force-dependent effective concentration decreases with applied force), which also explains why we observe blocking with soluble ACE2, even though the concentration of soluble ACE2 used is ∼10-fold lower (3.8 µM) than the effective tethered ACE2 concentration at zero force. Under force, the complex will still spend considerable time in the dissociated state, allowing free ACE2 from the solution to bind. Once soluble ACE2 is bound, it will be bound, on average, for τ0,bound ≈ 115 s, since the free ACE2 experiences no force, and therefore bias the system toward the open conformation.
With our tethered ligand assay, we investigated the interaction of an individual RBD:ACE2 interaction. On the virus membrane of SARS-CoV particles, however, spike proteins are trimeric, and multiple trimers are available for binding ACE2, which is known to cluster on the cell surface (3). Previous studies suggest that, at high loading rates, the force stability increases linearly with the number of interaction partners, while, at lower loading rates, a square root or logarithmic scaling is observed (66, 67). Multiple parallel interactions between RBDs and ACE2 thus could significantly strengthen the overall force stability, in particular at high loading rates. Our data suggest that, if held in close proximity, SARS-CoV-2 RBDs can engage ACE2 rapidly, within τ0,diss ≈ 70 ms. While our assay is different from the situation in vivo, the tethered ligand mimics the effect of preformed interactions by other RBDs on the same SARS-CoV-2 particle. This observation suggests that multivalent interactions between the virus and its host cell could form rapidly after an initial binding event, additionally providing increased stability of the interaction. We estimate the concentration of Spike in vivo as ∼1 pM, based on 7 ⋅ 106 viral copies in 1 mL sputum (8) and 100 Spike proteins per virus (1, 68). This estimated bulk protein concentration in vivo is much lower than the dissociation constants reported, which are in the range Kd ≈ 1 nM to 100 nM for the SARS-CoV-2 RBD binding to ACE2 and 10-fold higher for SARS-CoV-1 (SI Appendix, Table S4), suggesting that multivalency might be critical for efficient viral binding. The rapid binding of RBDs held in proximity to ACE2 revealed by our assay might, therefore, be an important component of SARS-CoV-2 infections. In addition, we can demonstrate and quantify blocking of the RBD:ACE2 bond. We anticipate that it will be a powerful tool to investigate the influence of mutations on interface stability and to assess the mode of action of potential therapeutic agents such as small molecules (69), neutralizing antibodies (58, 70), nanobodies (61, 71, 72), or designer proteins (73, 74) that interfere with S binding to ACE2. In particular, the tethered ligand assay could go beyond standard bulk assays and reveal heterogeneity, include avidity effects, investigate the ability of direct displacement, and determine drug residence times, in addition to affinities. Investigating how mutations in the RBD affect force stability might give valuable insights into reasons for the fitness advantages of newly emerging variants and might even provide a tool to predict those advantages based on increased force stability.
Materials and Methods
Engineering of Recombinant Proteins, Experimental Procedures, and Data Analysis.
SARS-CoV-1 and SARS-CoV-2 RBD:ACE2 tethered ligand constructs were designed based on available crystal structures PDB ID 2ajf (32) and PDB ID 6m0j (31). Please refer to SI Appendix for details of how tagged protein constructs were expressed, purified, and coupled to the surfaces for specific SMFS experiments. AFM and MT measurements were performed on custom-built instruments (34, 39) that were calibrated using the equipartition theorem to determine the spring constant of the AFM cantilever and the forces based on long DNA tethers in MT, respectively. The same SARS-CoV-1 and SARS-CoV-2 RBD:ACE2 constructs were investigated with both SMFS techniques at constant force-loading rate (AFM) and constant force (MT), respectively. Equilibrium measurements with MT and rupture experiments with an AFM were evaluated with custom MATLAB and Python scripts to deduce force stability and kinetics. SMD simulations were used to determine underlying molecular mechanisms. Details on the SMFS measurements, data analysis, and SMD simulations are given in SI Appendix.
Measurement Conditions.
Measurements of tethered ligand constructs in AFM and MT were performed in TBS (25 mM Tris, 72 mM NaCl, 1 mM CaCl2 at pH 7.2 and 22 °C). To test blocking, recombinant human ACE2 (GLN18-SER740, C-terminal His-tag) from RayBiotech (∼3.8 µM in measurement buffer, # 230-30165-100) was flushed into the flow cell and briefly incubated before the measurement.
Supplementary Material
Acknowledgments
We thank Meike Bos, Joost de Graf, David Dulin, and Klaus Überla for helpful discussions and Nina Beier, Benedikt Böck, and Ellis Durner for help with initial experiments. This study was supported by German Research Foundation Projects 386143268 and 111166240, a Human Frontier Science Program Cross Disciplinary Fellowship (LT000395/2020C) and European Molecular Biology Organization Non-Stipendiary long-term fellowship (ALTF 1047-2019) to L.F.M., and the Physics Department of LMU Munich. R.C.B. and P.S.F.C.G. are supported by start-up funds provided by Auburn University, and D.L. acknowledges support from the Spanish Ministry of Science, Innovation and Universities for the Spanish State Research Agency Retos Grant RTI2018-099318-B-I00, cofunded by the European Regional Development Fund.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2114397119/-/DCSupplemental.
Data Availability
The force spectroscopy dataset and MT and SMD datasets and analysis have been deposited in Figshare (https://figshare.com/articles/dataset/Atomic_force_microscopy_AFM_based_force_spectroscopy_dataset_for_the_SARS-CoV-2_RBD_ACE2_interaction/15062373).
References
- 1.Bar-On Y. M., Flamholz A., Phillips R., Milo R., SARS-CoV-2 (COVID-19) by the numbers. eLife 9, e57309 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li W., et al. , Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426, 450–454 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jia H. P., et al. , ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J. Virol. 79, 14614–14621 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou P., et al. , A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffmann M., et al. , SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181, 271–280.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shang J., et al. , Structural basis of receptor recognition by SARS-CoV-2. Nature 581, 221–224 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu M., et al. , Real-time conformational dynamics of SARS-CoV-2 spikes on virus particles. Cell Host Microbe 28, 880–891.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wölfel R., et al. , Virological assessment of hospitalized patients with COVID-2019. Nature 581, 465–469 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Lamers M. M., et al. , SARS-CoV-2 productively infects human gut enterocytes. Science 369, 50–54 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knowles M. R., Boucher R. C., Mucus clearance as a primary innate defense mechanism for mammalian airways. J. Clin. Invest. 109, 571–577 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rivas-Pardo J. A., Badilla C. L., Tapia-Rojo R., Alonso-Caballero Á., Fernández J. M., Molecular strategy for blocking isopeptide bond formation in nascent pilin proteins. Proc. Natl. Acad. Sci. U.S.A. 115, 9222–9227 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alsteens D., et al. , Nanomechanical mapping of first binding steps of a virus to animal cells. Nat. Nanotechnol. 12, 177–183 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Cuellar-Camacho J. L., et al. , Quantification of multivalent interactions between sialic acid and influenza A virus spike proteins by single-molecule force spectroscopy. J. Am. Chem. Soc. 142, 12181–12192 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Koehler M., Delguste M., Sieben C., Gillet L., Alsteens D., Initial step of virus entry: Virion binding to cell-surface glycans. Annu. Rev. Virol. 7, 143–165 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Florin E. L., Moy V. T., Gaub H. E., Adhesion forces between individual ligand-receptor pairs. Science 264, 415–417 (1994). [DOI] [PubMed] [Google Scholar]
- 16.Rief M., Gautel M., Oesterhelt F., Fernandez J. M., Gaub H. E., Reversible unfolding of individual titin immunoglobulin domains by AFM. Science 276, 1109–1112 (1997). [DOI] [PubMed] [Google Scholar]
- 17.Neuman K. C., Nagy A., Single-molecule force spectroscopy: Optical tweezers, magnetic tweezers and atomic force microscopy. Nat. Methods 5, 491–505 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Milles L. F., Schulten K., Gaub H. E., Bernardi R. C., Molecular mechanism of extreme mechanostability in a pathogen adhesin. Science 359, 1527–1533 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strick T. R., Allemand J. F., Bensimon D., Bensimon A., Croquette V., The elasticity of a single supercoiled DNA molecule. Science 271, 1835–1837 (1996). [DOI] [PubMed] [Google Scholar]
- 20.Lipfert J., Hao X., Dekker N. H., Quantitative modeling and optimization of magnetic tweezers. Biophys. J. 96, 5040–5049 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim J., Zhang C. Z., Zhang X., Springer T. A., A mechanically stabilized receptor-ligand flex-bond important in the vasculature. Nature 466, 992–995 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rognoni L., Stigler J., Pelz B., Ylänne J., Rief M., Dynamic force sensing of filamin revealed in single-molecule experiments. Proc. Natl. Acad. Sci. U.S.A. 109, 19679–19684 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kostrz D., et al. , A modular DNA scaffold to study protein-protein interactions at single-molecule resolution. Nat. Nanotechnol. 14, 988–993 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Le S., Yu M., Yan J., Direct single-molecule quantification reveals unexpectedly high mechanical stability of vinculin-talin/α-catenin linkages. Sci. Adv. 5, eaav2720 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Halvorsen K., Schaak D., Wong W. P., Nanoengineering a single-molecule mechanical switch using DNA self-assembly. Nanotechnology 22, 494005 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Kilchherr F., et al. , Single-molecule dissection of stacking forces in DNA. Science 353, aaf5508 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Yang D., Ward A., Halvorsen K., Wong W. P., Multiplexed single-molecule force spectroscopy using a centrifuge. Nat. Commun. 7, 11026 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jöhr R., Bauer M. S., Schendel L. C., Kluger C., Gaub H. E., Dronpa: A light-switchable fluorescent protein for opto-biomechanics. Nano Lett. 19, 3176–3181 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Shrestha P., et al. , Single-molecule mechanical fingerprinting with DNA nanoswitch calipers. Nat. Nanotechnol. 16, 1362–1370 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baumann F., et al. , Increasing evidence of mechanical force as a functional regulator in smooth muscle myosin light chain kinase. eLife 6, 621 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lan J., et al. , Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581, 215–220 (2020). [DOI] [PubMed] [Google Scholar]
- 32.Li F., Li W., Farzan M., Harrison S. C., Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 309, 1864–1868 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Ott W., et al. , Elastin-like polypeptide linkers for single-molecule force spectroscopy. ACS Nano 11, 6346–6354 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Löf A., et al. , Multiplexed protein force spectroscopy reveals equilibrium protein folding dynamics and the low-force response of von Willebrand factor. Proc. Natl. Acad. Sci. U.S.A. 116, 18798–18807 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huhle A., et al. , Camera-based three-dimensional real-time particle tracking at kHz rates and Ångström accuracy. Nat. Commun. 6, 5885 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dulin D., et al. , High spatiotemporal-resolution magnetic tweezers: Calibration and applications for DNA dynamics. Biophys. J. 109, 2113–2125 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walker P. U., Vanderlinden W., Lipfert J., Dynamics and energy landscape of DNA plectoneme nucleation. Phys. Rev. E 98, 042412 (2018). [Google Scholar]
- 38.Gumpp H., Stahl S. W., Strackharn M., Puchner E. M., Gaub H. E., Ultrastable combined atomic force and total internal fluorescence microscope. Rev. Sci. Instrum. 80, 063704 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Bauer M. S., et al. , Structural and mechanistic insights into mechanoactivation of focal adhesion kinase. Proc. Natl. Acad. Sci. U.S.A. 116, 6766–6774 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Puchner E. M., Franzen G., Gautel M., Gaub H. E., Comparing proteins by their unfolding pattern. Biophys. J. 95, 426–434 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hati S., Bhattacharyya S., Impact of thiol-disulfide balance on the binding of Covid-19 spike protein with angiotensin-converting enzyme 2 receptor. ACS Omega 5, 16292–16298 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ribeiro J. V., et al. , QwikMD - Integrative molecular dynamics toolkit for novices and experts. Sci. Rep. 6, 26536 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Phillips J. C., et al. , Scalable molecular dynamics on CPU and GPU architectures with NAMD. J. Chem. Phys. 153, 044130 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bustamante C., Marko J. F., Siggia E. D., Smith S., Entropic elasticity of lambda-phage DNA. Science 265, 1599–1600 (1994). [DOI] [PubMed] [Google Scholar]
- 45.Bouchiat C., et al. , Estimating the persistence length of a worm-like chain molecule from force-extension measurements. Biophys. J. 76, 409–413 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dietz H., Rief M., Exploring the energy landscape of GFP by single-molecule mechanical experiments. Proc. Natl. Acad. Sci. U.S.A. 101, 16192–16197 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bell G. I., Models for the specific adhesion of cells to cells. Science 200, 618–627 (1978). [DOI] [PubMed] [Google Scholar]
- 48.Verdorfer T., et al. , Combining in vitro and in silico single-molecule force spectroscopy to characterize and tune cellulosomal scaffoldin mechanics. J. Am. Chem. Soc. 139, 17841–17852 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laskowski R. A., et al. , PDBsum: A web-based database of summaries and analyses of all PDB structures. Trends Biochem. Sci. 22, 488–490 (1997). [DOI] [PubMed] [Google Scholar]
- 50.Humphrey W., Dalke A., Schulten K., VMD: Visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996). [DOI] [PubMed] [Google Scholar]
- 51.Webb B., Sali A., Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinformatics 54, 5.6.1–5.6.37 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ståhl S., et al. , Affibody molecules in biotechnological and medical applications. Trends Biotechnol. 35, 691–712 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Bernardi R. C., et al. , Mechanisms of nanonewton mechanostability in a protein complex revealed by molecular dynamics simulations and single-molecule force spectroscopy. J. Am. Chem. Soc. 141, 14752–14763 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Melo M. C. R., Bernardi R. C., de la Fuente-Nunez C., Luthey-Schulten Z., Generalized correlation-based dynamical network analysis: A new high-performance approach for identifying allosteric communications in molecular dynamics trajectories. J. Chem. Phys. 153, 134104 (2020). [DOI] [PubMed] [Google Scholar]
- 55.Yi C., et al. , Key residues of the receptor binding motif in the spike protein of SARS-CoV-2 that interact with ACE2 and neutralizing antibodies. Cell. Mol. Immunol. 17, 621–630 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moreira R. A., Chwastyk M., Baker J. L., Guzman H. V., Poma A. B., Quantitative determination of mechanical stability in the novel coronavirus spike protein. Nanoscale 12, 16409–16413 (2020). [DOI] [PubMed] [Google Scholar]
- 57.Cao W., et al. , Biomechanical characterization of SARS-CoV-2 spike RBD and human ACE2 protein-protein interaction. Biophys. J. 120, 1011–1019 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Starr T. N., et al. , Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell 182, 1295–1310.e20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walls A. C., et al. , Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181, 281–292.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Q., et al. , Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell 181, 894–904.e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wrapp D., et al. , VIB-CMB COVID-19 Response Team, Structural basis for potent neutralization of betacoronaviruses by single-domain camelid antibodies. Cell 181, 1004–1015.e15 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang J., et al. , Molecular interaction and inhibition of SARS-CoV-2 binding to the ACE2 receptor. Nat. Commun. 11, 4541 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sørensen C. S., Kjaergaard M., Effective concentrations enforced by intrinsically disordered linkers are governed by polymer physics. Proc. Natl. Acad. Sci. U.S.A. 116, 23124–23131 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.E. M. Mulhall et al., Single-molecule force spectroscopy reveals the dynamic strength of the hair-cell tip-link connection. Nat. Commun. 12, 849 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Krishnamurthy V. M., Semetey V., Bracher P. J., Shen N., Whitesides G. M., Dependence of effective molarity on linker length for an intramolecular protein-ligand system. J. Am. Chem. Soc. 129, 1312–1320 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Friddle R. W., Noy A., De Yoreo J. J., Interpreting the widespread nonlinear force spectra of intermolecular bonds. Proc. Natl. Acad. Sci. U.S.A. 109, 13573–13578 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Seifert U., Rupture of multiple parallel molecular bonds under dynamic loading. Phys. Rev. Lett. 84, 2750–2753 (2000). [DOI] [PubMed] [Google Scholar]
- 68.Ke Z., et al. , Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature 588, 498–502 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tiwari V., Beer J. C., Sankaranarayanan N. V., Swanson-Mungerson M., Desai U. R., Discovering small-molecule therapeutics against SARS-CoV-2. Drug Discov. Today 25, 1535–1544 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Baum A., et al. , Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science 369, 1014–1018 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huo J., et al. , Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2. Nat. Struct. Mol. Biol. 27, 846–854 (2020). [DOI] [PubMed] [Google Scholar]
- 72.Schoof M., et al. , QCRG Structural Biology Consortium, An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike. Science 370, 1473–1479 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chan K. K., et al. , Engineering human ACE2 to optimize binding to the spike protein of SARS coronavirus 2. Science 369, 1261–1265 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cao L., et al. , De novo design of picomolar SARS-CoV-2 miniprotein inhibitors. Science 370, 426–431 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The force spectroscopy dataset and MT and SMD datasets and analysis have been deposited in Figshare (https://figshare.com/articles/dataset/Atomic_force_microscopy_AFM_based_force_spectroscopy_dataset_for_the_SARS-CoV-2_RBD_ACE2_interaction/15062373).