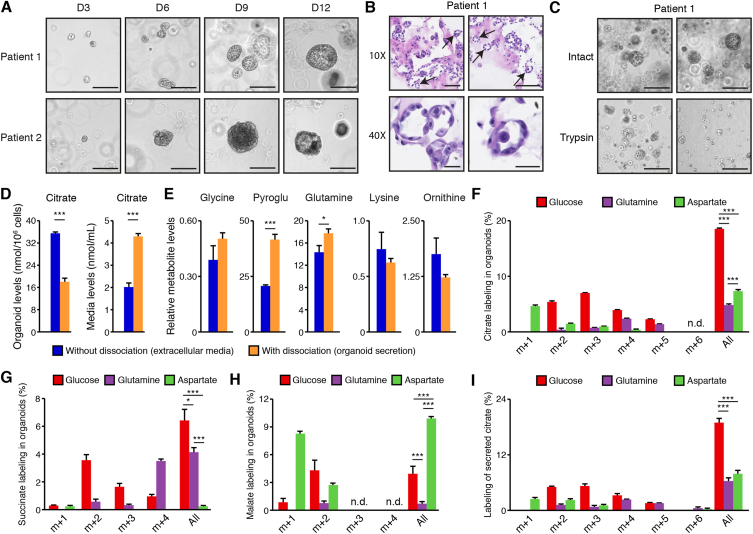
Figure 4.
Metabolomics analyses in human prostate organoids. A) Brightfield visualization of normal human prostate organoids over 12 days in three-dimensional culture. Organoid cultures derived from two different human patients are shown. Scale bars = 200 μm. B) H&E visualization of patient-derived prostate organoids showing the presence of internal lumen, demonstrated by black arrows. Scale bars = 125 μm and 50 μm, respectively, for 10× and 40× view. C) Brief treatment with trypsin (3 min) disrupts the three-dimensional architecture of human prostate organoids to allow connection of the internal lumen with the extracellular media. Scale bars = 200 μm. Representative images from patient #1 are shown and images for patient-derived organoids from patient #2 are available in Supplemental Figure S4C. D) Intra-organoid and extracellular citrate level quantification by GC–MS, with and without dissociation as shown in C. With dissociation, media contains luminal secretion. E) GC–MS metabolite quantification in extracellular culture media of human prostate organoids, with and without dissociation, as shown in C. For D and E, results are shown as the average ± SEM of one representative experiment performed in triplicate out of three independent experiments. Pyroglu: pyroglutamate. Stable isotope tracer analysis in organoids using 13C6-glucose, 13C5-glutamine, or 13C1-4-aspartate tracers is then shown for intra-organoid citrate (F), succinate (G), and malate (H) levels, as well as for secreted citrate levels (I). For results shown in F–I, organoids were dissociated before GC–MS metabolomics quantifications. Results are shown as the average ± SEM of one representative experiment performed in triplicates out of two independent experiments for patient #1. For these tracer experiments, note that organoids were cultured in parallel with the different 13C-labeled nutrients. Similar results were obtained with organoids from patient #2 and are shown in Supplemental Figure S4. N.d.: not detectable or below the limit of quantification. “All” indicates the sum of all shown isotopomers for a given metabolite. Statistics were only calculated using the sum of all isotopomers of a given metabolite. ∗∗∗p < 0.001; ∗∗p < 0.01; ∗p < 0.05.