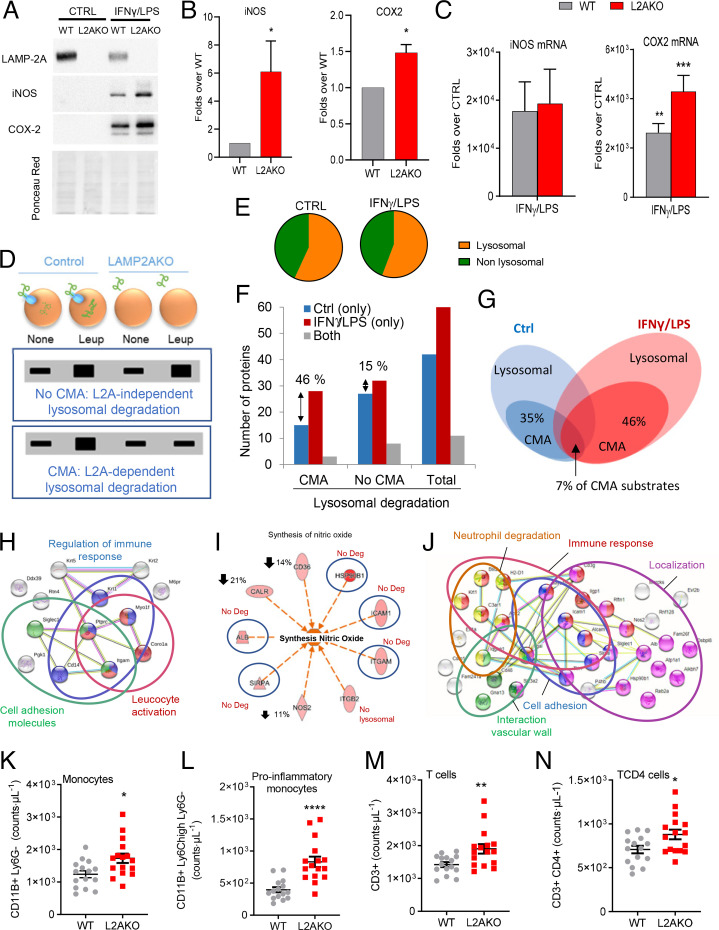
Fig. 4.
CMA blockage leads to an exacerbated proinflammatory phenotype in macrophages. (A and B) Levels of iNOS and COX2 proteins in BMDMs from WT and L2AKO mice cultured without additions (CTRL) or stimulated with IFNγ+LPS. Representative immunoblot (A) and densitometric quantification (B) expressed as folds over WT levels (n = 5 iNOS, n = 3 COX2). Ponceau red is shown as loading control. (C) mRNA levels of iNOS and Cox2 in the same cells expressed as folds over untreated (CTRL) (n = 4 iNOS, n = 3 Cox2). (D–J) Comparative proteomic analysis of lysosomes isolated from untreated (none) or leupeptin-treated WT and L2AKO BMDMs untreated (CTRL) or exposed to IFNγ+LPS from a pool of three individual experiments. Schematic of the experimental design and anticipated results for hypothetical proteins undergoing CMA-dependent or -independent lysosomal degradation (D). Percentage of lysosomal (constituents) and nonlysosomal proteins (substrates) in the fractions from CTRL and IFNγ+LPS macrophages (E). Number (F) and percentage (G) of proteins undergoing lysosomal degradation (Total) in a LAMP-2A–dependent (CMA) or -independent (no CMA) manner. STRING analysis for the top intracellular networks of CMA substrates in CTRL (H) and IFNγ+LPS (J) BMDMs. Detail of changes in degradation of proteins involved in synthesis of NO (I); blue circles indicate proteins no longer degraded in lysosomes in the L2AKO group, and down arrows indicate the reduction in lysosomal degradation of those proteins in the same group. (K–N) Number of total monocytes (K), proinflammatory subtype of monocytes (L), total T cells (M), and TCD4 cells (N) in WT and L2AKO mice (n = 15 WT, n = 16 L2AKO). All data, when applicable, were tested for normal distribution using the D’Agostino and Pearson normality test. Variables that did not pass the normality test were subsequently analyzed using the Mann–Whitney U rank-sum test. All other variables were tested with the Student’s t test. All values are mean ± SEM. Individual values are shown also in K–N. *P < 0.05, **P < 0.01, ***P < 0.005, and ****P < 0.001.