Fig. 2 |. Low R:FR exposure induces genome-wide binding of PIF7.
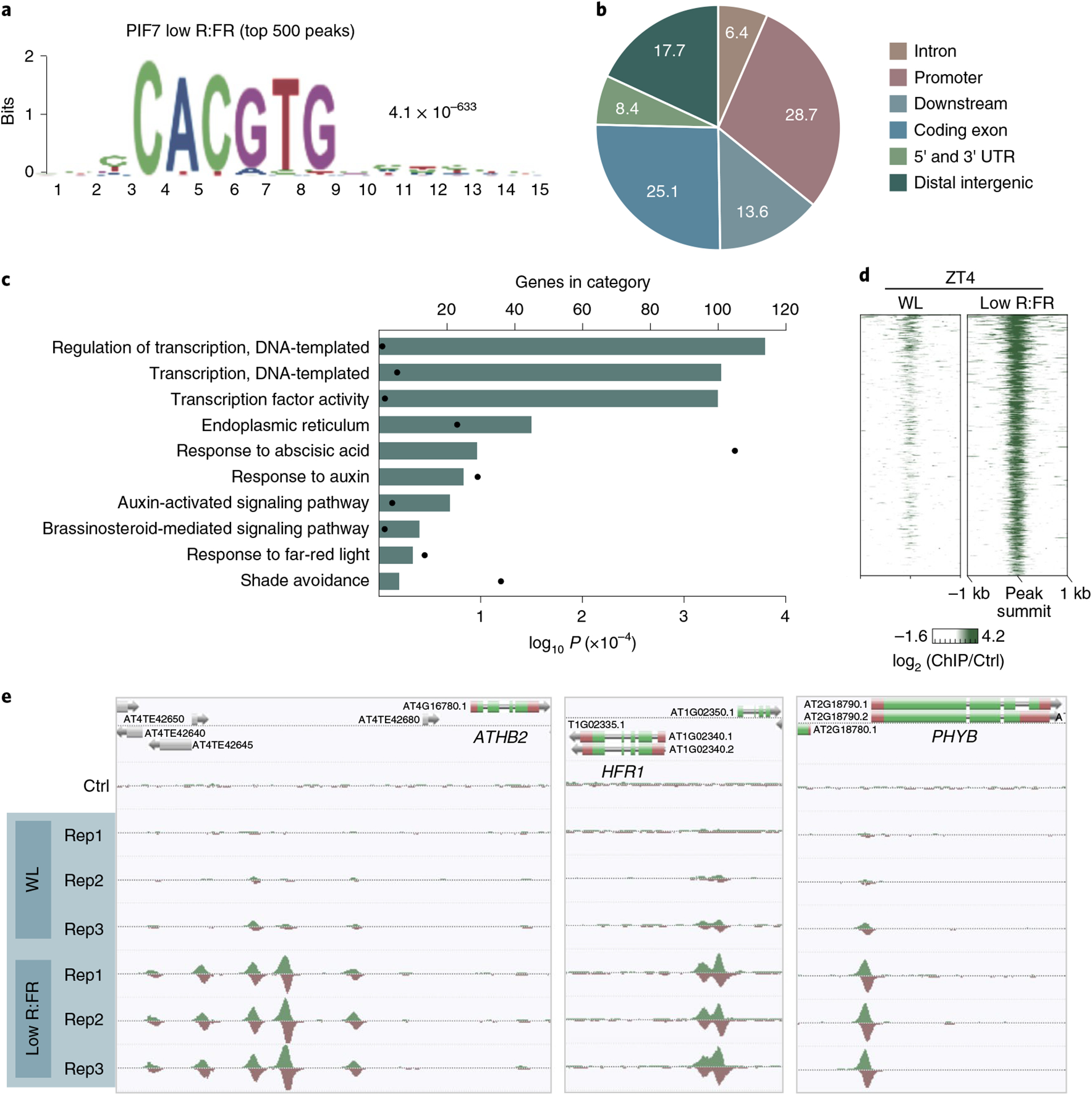
a, The CACGTG motif was the top-ranked motif in PIF7 ChIP–seq derived from low R:FR–exposed pif457 PIF7:PIF7:4xMYC seedlings at ZT4. Motif discovery was done through MEME analysis using the 500 top-ranked peaks. b, Pie chart illustrating the genomic distribution of PIF7-binding events after low R:FR exposure at ZT4. Numbers are proportions in percent. c, Gene ontology enrichment analysis reveals an enrichment for auxin signaling in the 500 top-ranked PIF7 targets. d, Heat map visualizing the low R:FR–induced DNA binding (log2 (fold change)) of PIF7 in pif457 PIF7:PIF7:4xMYC seedlings at ZT4. PIF7 DNA binding is shown for the top 500 binding sites that display the strongest low R:FR–induced PIF7 binding at ZT4 with each row of the heat map representing one binding event. kb, kilobase. e, AnnoJ genome browser screenshot visualizing PIF7 binding at ATHB2, HFR1 and PHYB. All tracks were normalized to the respective sequencing depth. Shown are three replicates (Rep1 to Rep3) per treatment. Ctrl, control.
