Figure 5.
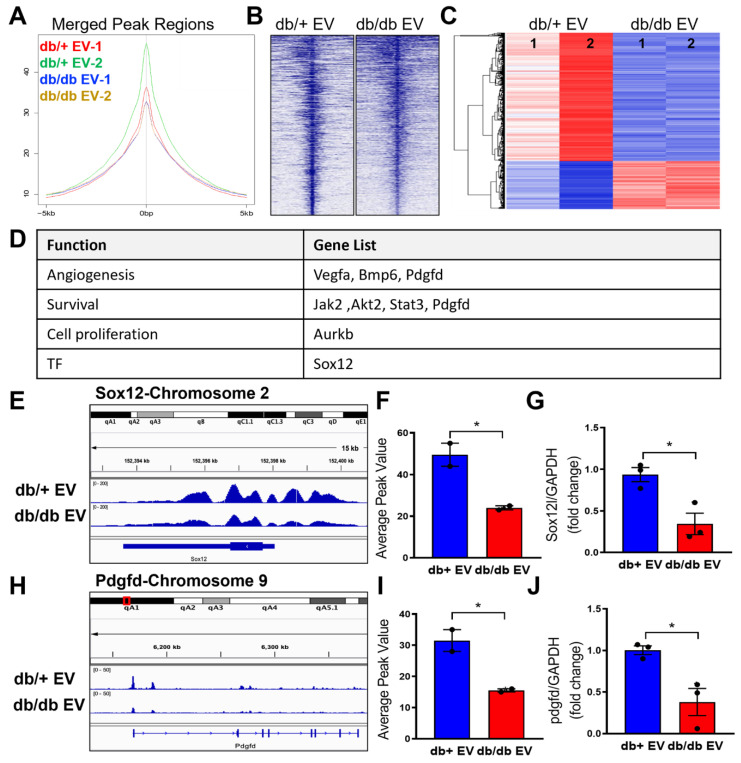
Non-diabetic EPC-EVs increase H3K9Ac level at the transcription start site of genes in MCECs. MCECs were treated with db/+ or db/db EPC-EVs for 24 hours and subjected to CHIP-seq against H3K9Ac. (A-B). db/db EPC-EVs decreased H3K9Ac level in MCECs. (A). Average peak values (y-axis) were plotted vs. sequencing tags across the TSS (-5000 to +5000 bp relative to the TSS; x-axis). (B). Sequencing tags distribution (y-axis) across the TSS (-5000 to +5000 bp relative to the TSS; x-axis) is presented as heatmaps. (C). The portion of differentially upregulated genes is greater in db/+ EPC-EV treated MCECs than db/db EPC-EV treated MCECs. Differentially expressed db/+ and db/db EPC-EV treated MCECs are shown as a heatmap in duplicate. n=2 for CHIP-seq experiment. (D). db/+ EPC-EV enhanced angiogenic/ cell survival/ proliferative gene expression. An upregulated gene list (FC > 2, P < 0.05) in functional categories. (E and H). db/db EPC-EV decreased H3K9Ac level at TSS of Sox12 and Pdgfd in MCEC. Integrative genomics viewer (IGV) software was used to view the fragment density of H3K9Ac (y-axis) aligned along with the gene coordinates (x-axis). (F and I) Data are presented as bar graphs representing the mean. n=2 for each group (G and J) db/db EPC-EV decreased Sox12 and Pdgfd expression in MCECs. Total RNA was extracted, Sox12 and Pdgfd gene expression were examined using qRT-PCR. The results were plotted. n=3 for qRT-PCR validation in each group. All Data are shown as mean ± SEM. *P<0.05. Pdgfd, platelet-derived growth factor D; Sox12, SRY-box transcription factor 12; TSS, transcription start site.