Significance
Estrogen receptor α (ERα) is a transcription factor that induces cell proliferation and exhibits increased expression in a large subset of breast cancers. We comprehensively searched for indicators of poor prognosis in ERα-positive breast cancer through the multiple databases, including interactome, transcriptome, and survival analysis, and identified FKBP52. We found that two immunophilins, FKBP52 and FKBP51, have opposing effects on ERα stability and propose that therapeutic targeting of FKBP52 could be useful for the prevention and treatment of ERα-positive breast cancers, including endocrine therapy–resistant breast cancers.
Keywords: BRCA1, breast cancer, estrogen receptor, FKBP, protein stability
Abstract
Estrogen receptor α (ERα) is a transcription factor that induces cell proliferation and exhibits increased expression in a large subset of breast cancers. The molecular mechanisms underlying the up-regulation of ERα activity, however, remain poorly understood. We identified FK506-binding protein 52 (FKBP52) as a factor associated with poor prognosis of individuals with ERα-positive breast cancer. We found that FKBP52 interacts with breast cancer susceptibility gene 1 and stabilizes ERα, and is essential for breast cancer cell proliferation. FKBP52 depletion resulted in decreased ERα expression and proliferation in breast cancer cell lines, including MCF7-derived fulvestrant resistance (MFR) cells, suggesting that inhibiting FKBP52 may provide a therapeutic effect for endocrine therapy–resistant breast cancer. In contrast, FKBP51, a closely related molecule to FKBP52, reduced the stability of ERα. Consistent with these findings, FKBP51 was more abundantly expressed in normal tissues than in cancer cells, suggesting that these FKBPs may function in the opposite direction. Collectively, our study shows that FKBP52 and FKBP51 regulate ERα stability in a reciprocal manner and reveals a regulatory mechanism by which the expression of ERα is controlled.
Estrogen receptor α (ERα), which is encoded by ESR1, is a transcriptional regulator that mediates developmental and physiological responses to the steroid hormone estrogen. ERα controls the expression of genes that regulate cell proliferation and differentiation in normal mammary tissue (1). The expression and activity of this receptor are increased in 70% of breast cancers (2). Proliferation of ERα-positive breast cancer cells is dependent on estrogen, and these cells respond to endocrine therapies which inhibit ERα signaling (3). However, a substantial number of ERα-positive breast cancers become resistant to endocrine therapy, which gives rise to recurrence, possibly as a result of increased protein stability of ERα (4, 5).
Up-regulation of ERα is mediated by several molecular mechanisms, including increased protein stability. Multiple proteins interact with ERα and protect it from polyubiquitylation-dependent proteolysis, leading to an accumulation of ERα at the protein level, thereby enhancing estrogen signaling and its physiological effects in breast cancer cells (6).
The ligand-unbound form of ERα exists as a monomer complexed with heat shock proteins (HSPs), mainly HSP70 and HSP90, and is distributed in both the nucleus and cytoplasm (7). Binding of ERα to estrogen triggers a major conformational change, which, in turn, triggers dissociation from HSPs, dimerization, and translocation into the nucleus (8). There, ERα recruits coactivators or corepressors to estrogen response elements (EREs) on the promoters of target genes, resulting in activation or repression of transcription, respectively (9). ERα undergoes polyubiquitylation mediated by multiple ubiquitin ligases (E3s) such as E6-associated protein (10), carboxyl terminus of HSC70-interacting protein (11), S-phase kinase–associated protein 2 (12), and murine double minute 2 (13), all of which induce proteasomal degradation (6). In contrast, other E3s, such as ring finger protein 8 (RNF8) (14), RNF31 (15), and Shank-associated RH domain interactor (SHARPIN) (16), interact with ERα and mediate monoubiquitylation of ERα, which prevents polyubiquitylation, thereby increasing its stability (6). Breast cancer susceptibility gene 1 (BRCA1) also mediates monoubiquitylation of ERα on lysines 302 and 303, which are also targeted by SHARPIN (17). Thus, multiple E3s contribute to intricate and elaborate regulation of ERα protein levels, as well as its transcriptional activity in breast cancer cells.
FK506-binding proteins (FKBPs) are a family of highly conserved proteins in eukaryotes. The prototype of this protein family is FKBP12 (18, 19), a binding partner for the immunosuppressive drugs FK506 and rapamycin (20). FKBP12 serves as a cis-trans peptidyl prolyl isomerase (PPIase), which catalyzes the interconversion between prolyl cis and trans conformations (21, 22). FKBPs contribute to diverse cellular functions, including protein folding, cellular signaling, apoptosis, and transcription (23). They function by directly binding to and altering the conformation of their target proteins, thereby acting as molecular switches. Of the 16 types of FKBPs in humans, FKBP51 and FKBP52, which are encoded by FKBP5 and FKBP4, respectively, act as regulators of nuclear receptors, including the glucocorticoid receptor (GR), progesterone receptor (PR), androgen receptor (AR), and ERα. FKBP52 positively regulates the function of these nuclear receptors (24–28), whereas FKBP51 negatively regulates the activity of GR (29) and PR (30), and positively regulates AR (28, 31). Both FKBPs have been implicated in the pathogenesis of prostate cancer (32, 33). Furthermore, FKBP5 expression is correlated with aggressiveness of cancers such as glioma and melanoma (34). However, elevated expression of FKBP4 is associated with tumor progression and poor prognosis in individuals with breast cancer (35). Although lines of evidence suggest that FKBP51 and FKBP52 affect cancer development and progression, the detailed mechanisms remain unknown.
In this study, we comprehensively searched for indicators of poor prognosis in ERα-positive breast cancer and identified FKBP52, which was found to be essential for breast cancer cell proliferation. Expression of FKBP52 was increased in ERα-positive breast cancer and stabilized ERα protein, whereas FKBP51 had the opposite effect. Collectively, our results uncover a regulatory mechanism underlying the expression of ERα.
Results
FKBP52 Expression Is Associated With Breast Cancer Prognosis.
To explore factors that affect the prognosis of individuals with ERα-positive breast cancer, we screened genes whose hazard ratio (HR) is calculable using the largest dataset of gene expression on Kaplan-Meier plotter (n = 12,180). We used the following four criteria: 1) genes with products that interact with ERα (n = 1,680); 2) genes whose messenger RNA (mRNA) expression correlates with expression of ERα mRNA, Pearson correlation coefficient > 0.2 (n = 962); 3) genes whose mRNA expression level is at least two times higher in the tumor than in the surrounding normal tissue (n = 749); and 4) genes whose mRNA expression level has an HR > 1 and log-rank P < 0.01 for the prognosis of ERα-positive breast cancer (n = 1,791) (Fig. 1A). Only three genes (FKBP4, CCND1, and GRHL2) met all of these criteria, with FKBP4 showing the highest score for HR. Given that CCND1 and GRHL2 have been extensively studied in association with breast cancer, we focused on FKBP4 and studied its biological effect on ERα as well as on the prognosis of individuals with ERα-positive breast cancer. Indeed, the expression of FKBP4 at the mRNA level was significantly higher in ERα-positive breast tumor tissue than in the surrounding normal tissue in The Cancer Genome Atlas (TCGA) database (Fig. 1B). Individuals with ERα-positive breast cancer displaying high FKBP4 expression had a poorer prognosis than those with low FKBP4 expression (Fig. 1C). These results suggest that the expression of FKBP4 is associated with the prognosis of patients with ERα-positive breast cancer.
Fig. 1.
High expression levels of FKBP52 correlate with poor prognosis in ER-positive breast cancer. (A) Isolation of factors closely related to ERα function were associated with poor prognosis in ERα-positive breast cancer. From a comprehensive database analysis, we isolated four groups of indicators of poor prognosis for breast cancer: factors that interact with ERα, factors with an HR of high expression significantly greater than 1, factors whose expression is significantly correlated (r > 0.2) with the expression of ESR1, and factors whose expression in ERα-positive breast cancer tissue is at least two times higher than that in the surrounding normal tissue. (B) The violin plot compares expression of FKBP4, which encodes FKBP52, in the surrounding normal tissue (green) and ERα-positive breast cancer tissue (in red) obtained from the TCGA BRCA database. P value was determined by Mann–Whitney U test. (C) FKBP4 mRNA level correlates with poor outcome in patients with ERα-positive (n = 3,082; Left), but not ERα-negative (n = 869; Right) breast cancer. The relapse-free survival plot based on expression of FKBP4 (200895_s_at) was derived from clinical cohorts acquired from the Kaplan-Meier Plotter database. Patients were distinguished using the median FKBP4 expression. HR was determined by two-tailed log-rank test. Correl, Pearson's correlation coefficient; FC, fold-change; norm, normalized count.
FKBP52 Stabilizes ERα Protein.
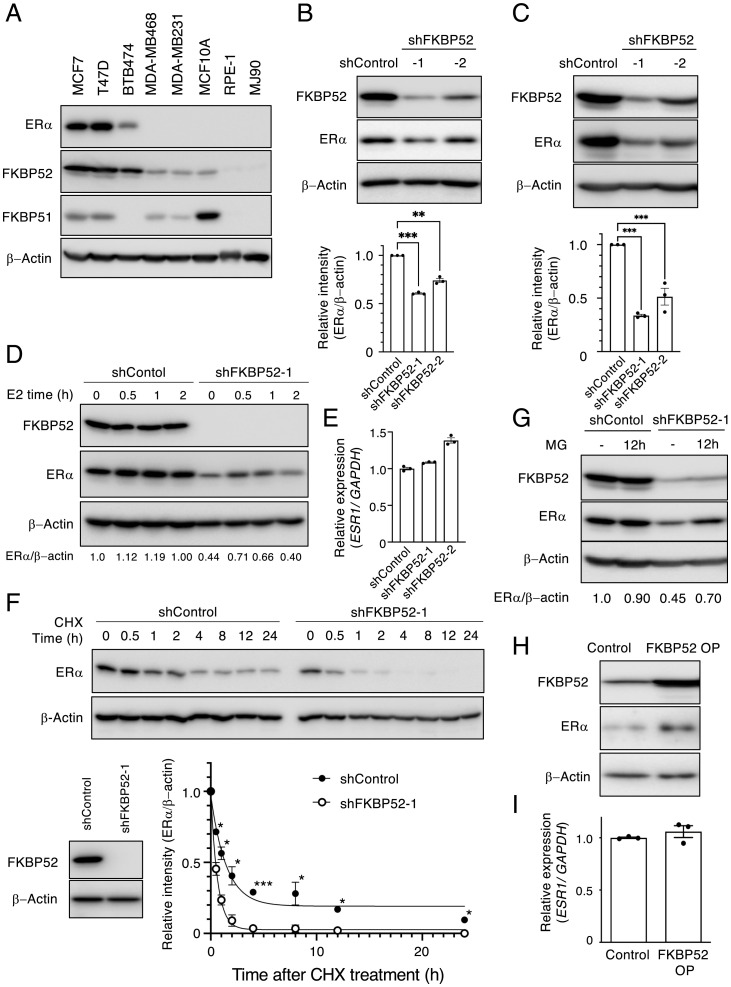
We examined the expression of the protein encoded by FKBP4 (FKBP52) by immunoblot analysis in various cell lines (Fig. 2A). The expression of FKBP52 was significantly higher in ERα-positive breast cancer cell lines (MCF7, T47D, and BTB474) than in ER−PR−HER2− breast cancer cell lines (MDA-MB468 and MDA-MB231), a normal mammary epithelial cell line (MCF10A), and other cell lines (RPE and MJ90). Depletion of FKBP52 mediated by two distinct short hairpin RNAs (shRNAs) in MCF7 cells resulted in a reduction in the abundance of total ERα at the protein level (Fig. 2B). We confirmed that total ERα was also reduced by FKBP52 depletion in T47D cells (Fig. 2C). Different molecular mechanisms have been suggested for the degradation (i.e., turnover) of ERα in the basal state (normal culture) and during E2 stimulation (36). Our results indicated that FKBP52 stabilizes ERα in the basal state and during E2 stimulation (Fig. 2D). In contrast, the expression of ESR1 encoding ERα at the mRNA level was not reduced by FKBP52 depletion (Fig. 2E), suggesting that the reduction in the protein level of ERα was likely attributable to posttranscriptional events. Thus, we examined whether the stability of the ERα protein was affected by FKBP52.
Fig. 2.
FKBP52 is involved in the stabilization of ERα. (A) FKBP52 expression levels in various cancer cells and normal cells. Total cell lysates were analyzed using immunoblotting with the indicated antibodies. (B and C) MCF7 (B) and T47D (C) cells expressing shControl or shFKBP52 were cultured in the presence of Dox for 2 d. Total cell lysates were analyzed using immunoblotting. The bar plot shows relative band intensities (mean ± SEM) of three independent experiments. **P < 0.01, ***P < 0.001 by Dunnett test. (D) MCF7 cells expressing shRNAs were cultured in medium containing charcoal-stripped serum and Dox for 2 d. The cells were collected after treatment with or without E2 for the indicated time and total cell lysates were analyzed using immunoblotting. The relative band intensity (ERα/β-actin) is mentioned at the bottom. (E) MCF7 cells expressing shRNAs were cultured in the presence of Dox for 3 d. Total RNA was prepared and the expression of ESR1 mRNA was determined using RT-qPCR. Results are shown as mean ± SEM of three independent experiments. (F) MCF7 cells expressing shRNAs were cultured in the presence of Dox for 1 d and then treated with 50 μg/mL cycloheximide (CHX) for the indicated time intervals. Total cell lysates were analyzed using immunoblotting. Results are expressed as the mean ± SEM of two independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 by one-tailed Student’s t test. (G) MCF7 cells expressing shRNAs were cultured in the presence or absence of proteasome inhibitor MG132 for 12 h. Total cell lysates were analyzed using immunoblotting. The relative band intensity (ERα/β-actin) is mentioned at the bottom. (H and I) MCF7 cells expressing FKBP52 or control were collected and subjected to immunoblotting (H), or the total RNA was prepared and expression of ESR1 mRNA was determined using RT-qPCR (I).
Cycloheximide chase experiments revealed that the stability of ERα protein was, indeed, diminished in MCF7 cells depleted of FKBP52 compared with that in control cells (Fig. 2F). The compromised stability of ERα was antagonized by treatment with the proteasome inhibitor MG-132 (Fig. 2G), suggesting that FKBP52 inhibits the ubiquitin-proteasome–mediated degradation of ERα. Consistent with this notion, overexpression of FKBP52 increased the abundance of ERα in MCF7 cells (Fig. 2H) without affecting its mRNA level (Fig. 2I).
FKBP52 Interacts with ERα.
We attempted to elucidate the mechanism of action underlying FKBP52-mediated stabilization of ERα. Lysates were prepared from MCF7 cells and subsequently immunoprecipitated with antibodies against FKBP52. The resultant immunoprecipitates were then subjected to immunoblot analysis using antibodies against ERα. This coimmunoprecipitation assay revealed that FKBP52 interacted with ERα in MCF7 cells (Fig. 3A). As observed in MCF7 cells, the interaction between endogenous FKBP52 and endogenous ERα was also detected in T47D cells (SI Appendix, Fig. S1A). ERα was localized in the nucleus in MCF7 cells that had been cultured in regular medium containing a physiological level of estrogens, and its distribution partially overlapped that of FKBP52 (Fig. 3B). FKBP52 depletion did not affect ERα localization (SI Appendix, Fig. S1B). Estrogen stimulation induces nuclear translocation and subsequent turnover of ERα. Subcellular fractionation of FKBP52-depleted MCF7 cells revealed that FKBP52 depletion did not affect the E2-induced dynamics of ERα (SI Appendix, Fig. S1C).
Fig. 3.
FKBP52 interacts with ERα. (A) The MCF7 cell lysates were prepared and FKBP52 was immunoprecipitated. The association of ERα and Hsp90 with FKBP52 were analyzed by immunoblotting. (B) Representative immunofluorescence image of MCF7 cells grown on coverslips, stained with the ERα antibody (green), FKBP52 (red), and DAPI (blue). (Scale bar, 10 μm.) (C) FKBP52 and FKBP51 contain several functional domains: peptidyl-prolyl cis/trans isomerase domain (FK1), the FK1-related domain (FK2), and the TPR domain. (D) Protein interactions between FKBP52 and ERα were detected using NanoBiT analysis in HEK293T cells. SmallBiT-FKBP52 (WT, F67D/D68V, K354A) and ERα-LargeBiT expression vectors were cotransfected into HEK293T cells. Cells were cultured in medium containing charcoal-stripped serum for 3 d. The bar graphs show the level of luminescence, which was caused by binding of FKBP52 to ERα 25 min after addition of the Nano-Glo Live Cell Reagent. The luminescence was measured with the luminometer (ARVO X4 [PerkinElmer]). Data are expressed as mean ± SEM of three independent experiments. *P < 0.05, **P < 0.01 by Dunnett test. (E) Effect of FK506 treatment on interaction between FKBP52 and ERα. Cells were treated with 50 μM FK506 for 2 h. The luminescence was measured after 5 min with the luminometer. Data are expressed as mean ± SEM of three independent experiments. ****P < 0.0001 by two-tailed Student’s t test. (F) The lysates of MCF7 cells stably expressing FKBP52 WT, F67D/D68V, or K354A were prepared, and FKBP52 was immunoprecipitated. The expression of ERα protein in FKBP52-overexpressing cells and the association of ERα and Hsp90 with FKBP52 were analyzed by immunoblotting. (G) Expression of ERα protein in FKBP52-overexpressing cells. MCF7 cells stably expressing FKBP52 WT, F67D/D68V, or K354A were cultured and prepared for immunoblotting. The relative ratios of ERα to β-actin are reported below the image. Ig, immunoglobulin; IP, immunoprecipitation.
FKBP52 and closely related FKBP51 contain two FKBP12-like domains (FK1 and FK2) and three tetratricopeptide repeats (TPRs) (Fig. 3C). PPIase activity and FK506 binding are restricted to FK1 (37). The interaction between FKBP52 and ERα was examined using a split luciferase assay (NanoBiT), and the interaction between FKBP52 (wild type [WT]) and ERα was confirmed (Fig. 3D). This interaction was abrogated by a mutation in the TPR domain (K354A), which is unable to interact with Hsp90, or by treatment with FK506, but not by PPIase-deficient mutations, in the FK1 domain (F67D/D68V) (Fig. 3 D and E). Consistent with the NanoBiT results, immunoprecipitation assays revealed that the FKBP52 WT and F67D/D68V mutant were associated with ERα, whereas the K354A mutant had a reduced ability to bind to ERα (Fig. 3F). Stabilization of ERα by overexpression of FKBP52 was impaired by either the F67D/D68V or K354A mutation (Fig. 3G). Collectively, we concluded that both the interaction of FKBP52 with ERα and its PPIase activity are necessary for the stabilization of ERα.
FKBP52 Up-Regulates Transcriptional Activity of ERα.
Next, we examined whether FKBP52 affected the transactivation function of ERα. Depletion of FKBP52 by shRNA-mediated RNA interference in MCF7 cells markedly inhibited cell proliferation (Fig. 4A). In FKBP52-depleted T47D (ERα-positive breast cancer cell line) and Hs578T (triple-negative breast cancer cell line) cells, proliferation was substantially attenuated by FKBP52 depletion, as observed in MCF7 cells (SI Appendix, Fig. S2 A and B). These results suggest that FKBP52 may also regulate cell proliferation in an ERα-independent manner. RNA-sequencing (RNA-seq) analysis was performed to comprehensively analyze the effects of FKBP depletion. The effect of shRNAs was clearly observed by hierarchical clustering and principal component analysis using RNA-seq data (SI Appendix, Fig. S3 A and B). Gene set enrichment analysis (GSEA) revealed that the sets of genes down-regulated and up-regulated in response to E2 exposure were positively and negatively enriched, respectively, in FKBP52-depleted cells (Fig. 4B). Ingenuity Pathway Analysis upstream regulator analysis was performed to predict upstream molecules that were down-regulated in FKBP52-depleted cells. ESR1 encoding ERα was found to be down-regulated, with the lowest P value (P = 4.29 × 10−36) in FKBP52-depleted cells (SI Appendix, Fig. S2C). These results suggest that the depletion of FKBP52 suppresses the transactivation of ERα. Depletion of FKBP52 in T47D cells attenuated the activity of luciferase fused with EREs in the presence of E2, compared with that in control cells (Fig. 4C). Treatment of cells with FK506 also showed concentration-dependent inhibition of luciferase activity in both T47D (Fig. 4D) and MCF7 cells (SI Appendix, Fig. S2D). Overexpression of WT FKBP52, but not the K354A mutant, in MCF7 cells enhanced luciferase activity in the presence of E2 compared with control cells (Fig. 4E), suggesting that the interaction is required for ERα regulation. On the other hand, decreased luciferase activity in the F67D/D68V overexpression mutants might reflect the dominant-negative effect of these mutations.
Fig. 4.
FKBP52 is involved in transcriptional activation of ERα target genes. (A) MCF7 cells expressing shControl or shFKBP52 were cultured in the presence of Dox, and cells were collected and counted. Data are expressed as the mean ± SEM of three independent experiments. *P < 0.05, ****P < 0.0001 by two-tailed Student’s t test. Knockdown efficiency by immunoblotting is shown below the graph. (B) GSEA profiles of estradiol response comparing MCF7 cells expressing shControl and shFKBP52. Sizes, NES (normalized enrichment score), nP (nominal P value), and FDR (false discovery rate) are shown. Three biological replicates were analyzed. (C) T47D cells expressing indicated shRNAs were cultured in medium containing charcoal-stripped serum and Dox for 1 d. The cells were transiently transfected with ERE-luciferase reporter plasmids. After 24 h of transfection, the cells were treated with E2 for another 24 h and luciferase activity was measured. Data are expressed as the mean ± SEM of three independent experiments. **P < 0.01 by two-tailed Student’s t test. (D) T47D cells were cultured in medium containing charcoal-stripped serum for 1 d. The cells were transiently transfected with ERE-luciferase reporter plasmids. After 48 h of transfection, the cells were treated with the indicated concentration of FK506. After 1 h of FK506 treatment, the cells were treated with 10 nM E2 for another 6 h and luciferase activity was measured. Data are expressed as the mean ± SEM of three independent experiments. **P < 0.01, ****P < 0.0001 by Dunnett test. (E) MCF7 cells stably expressing FKBP52 WT, F67D/D68V, or K354A were cultured in medium containing charcoal-stripped serum for 1 d. The cells were transiently transfected with ERE-luciferase reporter plasmids. After 24 h of transfection, the cells were treated with E2 for another 24 h and luciferase activity was measured. The results from three independent experiments are shown. Data are expressed as the mean ± SEM *P < 0.05 by Dunnett test. (F) MCF7 cells expressing shControl or shFKBP52 were transplanted subcutaneously into the flank of NOD/Shi-scid IL-2RγKO mice (n = 7). Tumor size was measured twice weekly. If tumor was undetected, tumor size was considered to be 0. *P < 0.05 by one-tailed Mann–Whitney U test. (G and H) Paraffin-embedded inoculated tumors arising from MCF7 cells expressing shControl (n = 4) or shFKBP52-1 (n = 2) were subjected to immunohistochemical assay using anti–Ki-67 antibody. Representative images of immunohistochemical staining for Ki-67 were obtained using NanoZoomer 2.0RS (G). (Scale bar, 20 μm.) The percentage of Ki-67–positive cells was determined using QuPath, version 0.3.1, software (H). *P < 0.05 by Student’s t test. DMSO, dimethyl sulfoxide.
Contrary to our results obtained using MCF7 cells, FKBP52 overexpression has been reported to show a tendency to decrease ERα signaling in SK-N-MC cells (a neuroblastoma cell line) (38). To verify whether these differences were caused by cell lines, a luciferase reporter assay using SK-N-MC cells was performed. Similar to a previous report (38), FKBP52 overexpression in SK-N-MC cells decreased ERα signaling (SI Appendix, Fig. S2E). These results suggest that the enhancement of ERα transcriptional activity by FKBP52 is specific to ERα-positive breast cancer cells.
Next, we used a mouse xenograft model to examine the tumor-suppressive effect of FKBP52 depletion in vivo. shRNA-mediated FKBP52-depleted or control MCF7 cells were subcutaneously transplanted into the flank of immunodeficient NOD/Shi-scid IL-2RγKO mice with E2 pellet supplementation, and the mice then were ovariectomized. The inoculated tumors arising from FKBP52-depleted MCF7 cells were either undetectable or considerably smaller than those arising from the controls at 70 and 77 d posttransplantation (Fig. 4F). Immunohistochemical analysis revealed the percentage of Ki-67–positive cells, a marker of cell proliferation, was lower in tumors arising from FKBP52-depleted cells than that from control cells (Fig. 4 G and H). Consistent with the in vitro results, these results suggest that FKBP52 is essential for tumor growth in vivo. These results also suggest that FKBP52 enhances ERα transcriptional activity and cell proliferation in ERα-positive breast cancer cells.
FKBP52 Inhibition Decreases Cell Proliferation in Fulvestrant-Resistant Breast Cancer Cells.
Resistance to endocrine therapy is a major limitation in ERα-positive breast cancer treatment. Therefore, the role of FKBP52 in endocrine therapy–resistant breast cancer cells was examined. Fulvestrant or FK506 alone reduced ERα expression and cell proliferation, and their combination further reduced ERα expression (Fig. 5A) and cell proliferation (Fig. 5B), suggesting that the combination of fulvestrant and FKBP52 inhibition is effective in ERα-positive breast cancer treatment. Next, we examined the effect of FKBP52 inhibition on ERα expression and cell proliferation in MCF7-derived fulvestrant resistance (MFR) cells (39). FK506 treatment decreased total ERα expression and reduced proliferation in MFR cells (Fig. 5 C and D). As seen in MCF7 and T47D cells, we confirmed decreased ERα expression and reduced cell proliferation in FKBP52-depleted MFR cells by shRNA (Fig. 5 E and F). These results suggest that FKBP52 inhibition had an additive effect on the decrease in ERα stability by fulvestrant, and FKBP52 inhibition also exerted tumor suppressive effects in fulvestrant-resistant cells.
Fig. 5.
FKBP52 depletion or inhibition decreases ERα expression and cell growth in fulvestrant (FULV)-resistant breast cancer cells. (A) MCF7 cells were treated with 25 or 50 μM FK506, and/or 100 nM FULV for 24 h. Total cell lysates were analyzed by immunoblotting. The relative ratios of ERα to β-actin are reported below the image. (B) MCF7 cells were cultured with 25 μM FK506 and/or 100 nM FULV for the indicated number of days. The cells were collected and counted. Data are expressed as the mean ± SEM of three independent experiments. ****P < 0.0001 by Dunnett t test. (C) MFR cells were treated with 50 μM FK506 for 24 h. Total cell lysates were analyzed by immunoblotting. The bar plot shows relative band intensities (mean ± SEM) of three independent experiments. ***P < 0.001, by two-tailed paired t test. (D) MFR cells were cultured with 25 μM FK506, and the cells were collected and counted. Data are expressed as the mean ± SEM of three independent experiments. ***P < 0.001, ****P < 0.0001 by two-tailed Student’s t test. (E) MFR cells expressing shControl or shFKBP52 were cultured with Dox for 3 d. Total cell lysates were analyzed by immunoblotting with the indicated antibodies. The bar plot shows relative band intensities (mean ± SEM) of three independent experiments. *P < 0.05, by two-tailed paired t test. (F) MFR cells expressing shControl or shFKBP52 were cultured with Dox, and the cells were collected and counted. Data are expressed as the mean ± SEM of three independent experiments. ****P < 0.0001 by two-tailed Student’s t test. DMSO, dimethyl sulfoxide.
FKBP51 and FKBP52 Have Opposing Effects on ERα.
FKBP51 is a member of the FKBP family and is most closely related to FKBP52. We examined the role of FKBP51 in the regulation of ERα. Depletion of FKBP51 resulted in an increase in the abundance of ERα to a modest extent in MCF7 breast cancer cells (Fig. 6A), whereas the effect was much more prominent in MCF10A normal mammary epithelial cells (Fig. 6B). Additionally, we found that the expression of ESR1, encoding ERα, was not greatly affected by FKBP51 depletion in both MCF7 (Fig. 6C) and MCF10A cells (SI Appendix, Fig. S4A), suggesting that the increased amount of ERα is likely attributable to posttranscriptional events in FKBP51-depleted cells. In MCF7 cells, cycloheximide chase experiments revealed that the half-life of ERα was modestly extended by FKBP51 depletion (Fig. 6D). FKBP51 depletion also enhanced the expression of ERα in response to E2 treatment (Fig. 6E).
Fig. 6.
Knockdown of FKBP51 increases the expression and transcriptional activity of ERα. (A and B) MCF7 (A) or MCF10A (B) cells expressing shControl or shFKBP51 were cultured in the presence of Dox for 2 d. Cells were collected and immunoblotting was performed using the indicated antibodies. The bar plot shows relative band intensities (mean ± SEM) of three independent experiments. *P < 0.05, **P < 0.01 by Dunnett t test (A) and two-tailed paired t test (B). (C) RT-qPCR analysis of ESR1 was performed in MCF7 cells expressing indicated shRNAs. Data are expressed as the mean ± SEM of three independent experiments. (D) MCF7 cells expressing indicated shRNAs were cultured and analyzed as shown in Fig. 2F. (E) MCF7 cells expressing indicated shRNAs were cultured and analyzed, as shown in Fig. 2D. The relative ratios of ERα to β-actin are reported below the image. (F) The lysates of MCF7 cells stably expressing 3× FLAG-tagged FKBP51 WT, F67D/D68V, or K352A/R356A were prepared and FLAG was immunoprecipitated. The association of ERα with FKBP51 was analyzed by immunoblotting. (G) T47D cells expressing indicated shRNAs were cultured in medium containing charcoal-stripped serum and Dox for 1 d. The cells were transiently transfected with ERE-luciferase reporter plasmids. After 24 h of transfection, the cells were treated with 10 nM E2 for another 24 h and luciferase activity was measured. Data are expressed as the mean ± SEM of three independent experiments. **P < 0.01 by two-tailed Student’s t test. CHX, cycloheximide; DMSO, dimethyl sulfoxide; IP, immunoprecipitation.
The interaction between FKBP51 and ERα was examined using immunoprecipitation, and it was determined that FKBP51 (WT) and FKBP51 (F67D/D68V) directly interacted with ERα (Fig. 6F). Additionally, this interaction was abrogated by FKBP51 (K352A/R356A). This abrogation was also observed in the NanoBiT assay (SI Appendix, Fig. S4B), suggesting that the TPR domain of FKBP51 is important for its interaction with ERα, as seen in FKBP52.
Consistent with increased ERα expression, FKBP51 depletion in T47D cells enhanced the activity of luciferase fused with EREs in the presence of E2 compared with that in control cells (Fig. 6G). GSEA revealed that the ERα target gene sets that were down-regulated and up-regulated in response to E2 exposure were negatively and positively enriched, respectively, in FKBP51-depleted MCF7 cells (SI Appendix, Fig. S4C), suggesting that FKBP51 depletion enhanced the transcriptional activity of ERα. Together, these results suggest that the action of FKBP51 on ERα stability and function is opposite to that of FKBP52.
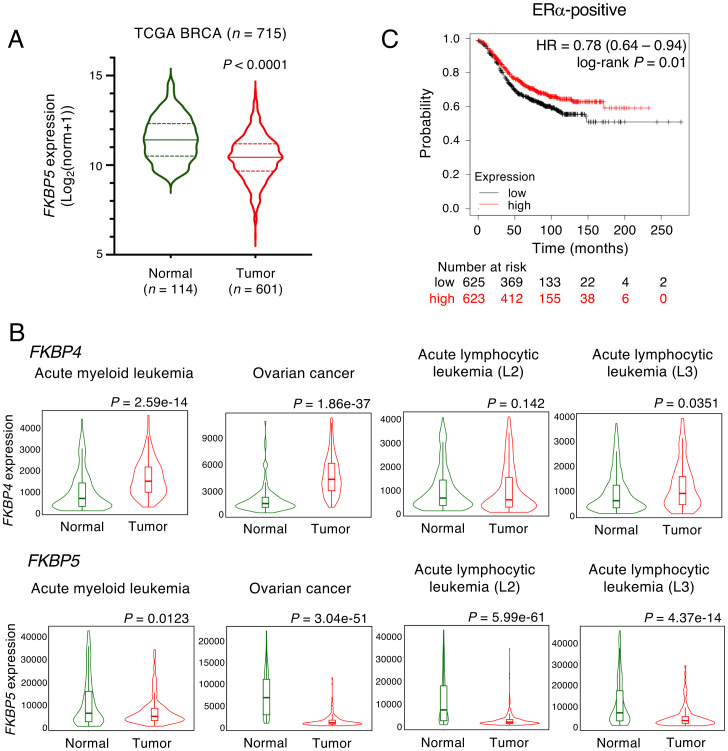
In contrast to FKBP4, the expression of FKBP5, encoding FKBP51, at the mRNA level was significantly lower in ERα-positive breast tumor tissue than in the surrounding normal tissue in the TCGA database (Fig. 7A). This opposing expression pattern of FKBP4 and FKBP5 in normal and tumor tissues was not restricted to hormone-sensitive breast cancer; it was also observed globally in several cancers (namely, acute myeloid leukemia, ovarian cancer, acute lymphocytic leukemia L2 and L3 subtypes) with significantly higher expression levels of ESR1 than in the surrounding normal tissues in the TNMplot database (40) (Fig. 7B). Furthermore, individuals with ERα-positive breast cancer with high FKBP5 expression had a better prognosis than those with low FKBP5 expression (Fig. 7C). Collectively, FKBP52 interacts with and thereby stabilizes ERα, resulting in the transcriptional activation of ERα target genes, whereas FKBP51 antagonizes the action of FKBP52 on ERα.
Fig. 7.
FKBP5 expression is low, while FKBP4 expression is high in tumors. (A) The violin plot compares expression of FKBP5, which encodes FKBP51, in normal surrounding tissue (green) and ERα-positive breast cancer tissue (red) obtained from the TCGA BRCA database. The P value was determined by Mann–Whitney U test. (B) Comparison of FKBP4 and FKBP5 expression in various normal and tumor tissues in which ESR1 is significantly overexpressed in tumor compared with normal samples from the TNMplot database (40). The P value was determined by Mann–Whitney U test. (C) Low expression of FKBP5 mRNA correlates with poor outcome in patients with ERα-positive breast cancer (n = 1,248). The relapse-free survival plot based on expression of FKBP5 (224856_at) was derived from clinical cohorts acquired from the Kaplan–Meier Plotter database. Patients were distinguished by median FKBP5 expression. HR was determined by two-tailed log-rank test. norm, normalized count.
BRCA1 Interacts with FKBP52 and Contributes to Increased ERα Stability.
We investigated the molecular mechanism that accounts for the opposing effects of FKBP52 and FKBP51 on the stability of ERα. We hypothesized that these FKBPs might affect the association of E3s with ERα. To explore this, we screened genes using the following three criteria: 1) genes with products that interact with ERα (n = 2,210), 2) genes with products that interact with FKBP52 (n = 125), and 3) genes encoding E3s (n = 377) (41) (Fig. 8A). Only one gene (BRCA1) met all these criteria.
Fig. 8.
BRCA1 is more highly expressed in ERα-positive tumor and mediates ERα stability together with FKBP52. (A) Identification of factors that interact with FKBP52 to regulate the stability of ERα. Using a comprehensive database, we analyzed E3 ligases that interact with both ERα and FKBP52 and obtained only one candidate, BRCA1. (B and C) MCF7 (B) and T47D (C) cells expressing shControl or shBRCA1 were cultured in the presence of Dox for 2 d. Cells were collected and immunoblotting was performed using the indicated antibodies. The bar plot shows relative band intensities (mean ± SEM) of three independent experiments. ***P < 0.001, ****P < 0.0001 by two-tailed paired t test (B) and Dunnett test (C). (D) MCF7 cells expressing shControl or shBRCA1 were cultured in the presence of Dox for 2 d. Cells were collected to extract total RNA and expression of ESR1 mRNA was determined using RT-qPCR. Data are expressed as the mean ± SEM of three independent experiments. (E) MCF7 cells expressing indicated shRNAs were cultured and analyzed as shown in Fig. 2F. (F and G) MCF7 cell lysates were prepared, and FKBP51, FKBP52 (F), or BRCA1 (G) was immunoprecipitated. The immunoprecipitants were analyzed by immunoblotting. (H) Scatterplot of ESR1 and BRCA1 expression in breast tumor tissues. BRCA1 expression levels were plotted in breast cancers with ESR1 expression levels of 10 or higher. (I) The violin plot compares expression of BRCA1 in normal surrounding tissue (green) and ERα-positive breast cancer tissue (red) generated from the TCGA BRCA database. The P value was determined by Mann–Whitney U test. (J) Model of estrogen-dependent transcriptional regulation mediated through FKBP51- and FKBP52-dependent ERα stability. FKBP52-bound ERα interacts with BRCA1, which monoubiquitylates ERα and protects it from degradation. In contrast, FKBP51 competes with FKBP52 for binding to ERα and destabilizes the protein. CHX, cycloheximide; Ig, immunoglobulin; IP, immunoprecipitation; norm, normalized count.
We found that depletion of BRCA1 in MCF7 and T47D cells resulted in a reduction in the abundance of ERα at the protein level (Fig. 8 B and C and SI Appendix, Fig. S5A) but not at the mRNA level (Fig. 8D). Cycloheximide chase experiments revealed that the stability of ERα protein was decreased in MCF7 cells depleted of BRCA1 compared with that in control cells (Fig. 8E and SI Appendix, Fig. S5B). Depletion of both FKBP52 and BRCA1 had little additive effect on the decrease in ERα stability induced by a single depletion of FKBP52 (SI Appendix, Fig. S5C), suggesting that FKBP52 and BRCA1 operate through the same pathway to regulate ERα stability. Coimmunoprecipitation analysis showed that BRCA1 interacted with FKBP52 but not with FKBP51 (Fig. 8 F and G).
To examine the role of each component in the formation of the FKBP52–BRCA1–ERα trimeric complex, a series of immunoprecipitation experiments were performed using MCF7 cells depleted of FKBP52 (SI Appendix, Fig. S5D), BRCA1 (SI Appendix, Fig. S5E), or ERα (SI Appendix, Fig. S5F). In FKBP52-depleted cells, the interaction between ERα and BRCA1 was significantly attenuated (SI Appendix, Fig. S5D), which is consistent with our model that FKBP52 promotes the binding of BRCA1 to ERα. Similarly, BRCA1 depletion reduced the interaction between FKBP52 and ERα (SI Appendix, Fig. S5E). In contrast, ERα knockdown did not affect the interaction between FKBP52 and BRCA1 (SI Appendix, Fig. S5F), suggesting that ERα did not contribute to the FKBP52-BRCA1 association. The mRNA expression of BRCA1 was positively correlated with that of ESR1 in the TCGA dataset (Fig. 8H). Furthermore, the abundance of BRCA1 mRNA was greater in the tumor than in the normal surrounding tissue (Fig. 8I). Given that BRCA1 mediates monoubiquitylation of ERα (17), which might prevent its polyubiquitylation and subsequent proteasomal degradation, these results suggest that FKBP52 recruits BRCA1 to ERα, thereby stabilizing it (Fig. 8J). FKBP51, which is unable to interact with BRCA1, might competitively antagonize this effect of FKBP52, resulting in increased stability of ERα.
Discussion
Although many nuclear receptors have been shown to be regulated by FKBPs, it is unclear how ERα is affected by FKBPs. In the present study, we identified FKBP4 as a candidate that affects the prognosis of individuals with ERα-positive breast cancer. The other candidates that met our criteria listed in Results were CCND1 and GRHL2, both of which have been well studied in the context of breast cancer. CCND1 encodes cyclin D1, which associates with and activates CDK4 and CDK6, promoting cell cycle progression from G0 to G1 phase (42). Overexpression of this gene is broadly observed in various types of cancer (43). Furthermore, CCND1 overexpression is associated with poor prognosis in patients with breast cancer (44). Inhibitors of CDK4 and CDK6 have been used in the treatment of breast cancer (45). GRHL2 encodes a transcription factor that has been shown to inhibit apoptosis, promote proliferation, and correlate with ESR1 expression in breast cancer (46–48). GRHL2 is recruited to target DNA by ERα to regulate the expression of estrogen-responsive genes (49, 50). The fact that these genes were identified as candidates through our screening confirms that our screening strategy was appropriate. Therefore, we assumed that FKBP4, like CCND1 and GRHL2, may contribute to the regulation of ERα and correlate with the prognosis of breast cancer. Thus, we investigated how FKBP52, encoded by FKBP4, affects the expression and function of ERα in relation to breast cancer.
FKBP52 has been shown to act as a positive regulator of several nuclear receptors, including GR, PR, and AR, by promoting their activity, stability, and nuclear translocation (24–28). However, little has been reported on the regulation of ERα by FKBP52. In the present study, we showed that FKBP52 binds to and stabilizes ERα, resulting in an increase in the abundance and activity of ERα. Furthermore, increased expression of FKBP52 is observed in ERα-positive breast cancer and is associated with poor patient prognosis. Depletion of FKBP52 leads to a reduction in ERα expression and compromised proliferation of breast cancer cells, which is consistent with the previous observation that ERα is essential for ERα-positive breast cancer cell proliferation (51). The tumor-suppressive effects of FKBP52 depletion were also observed in fulvestrant-resistant breast cancer cells, suggesting that the pharmacological inhibition of FKBP52 may be a promising approach to address endocrine-therapy resistance. Given that a reduction in the transcriptional activity of ERα was observed in FKBP52-deficient mouse embryonic fibroblast, FKBP52 may regulate ERα in vivo (27). In contrast, FKBP51, a member of the FKBP family that is most closely related to FKBP52, also binds to ERα; however, it reduces the stability of ERα, resulting in the suppression of its function. Of note, FKBP51 depletion increased ERα expression in MCF10A cells, which is thought to be an ERα-negative cell line. Re-expression of ERα in ERα-negative breast cancer cells restores sensitivity to endocrine therapy (52). FKBP51 inhibition may contribute to this notion.
We explored E3s that might interact with both FKBP52 and ERα, using the comprehensive interactome database, and identified only BRCA1. Thus, we speculate that FKBP52 might promote the binding of BRCA1 to ERα and contribute to its stabilization. However, the effect of BRCA1 on ERα transcriptional activity remains controversial. A previous study showed that BRCA1 promoted the degradation of ERα protein (53), whereas it was required for ERα expression at the transcriptional level (54). In contrast, our results showed that the stability of ERα was, indeed, reduced in cells depleted of BRCA1, suggesting that BRCA1 contributes to increased ERα stability. Although the basis for the inconsistency between these two studies is currently unclear, the effect of BRCA1 on ERα might be dependent on the cellular context. Of note, the mRNA expression of BRCA1 and ESR1 was correlated (r = 0.2583) and BRCA1 was highly expressed in ERα-positive breast cancer tissues compared with normal breast tissue. These findings support the notion that FKBP52 interacts with ERα and recruits BRCA1, which mediates monoubiquitylation of ERα, thereby protecting ERα from proteasomal degradation (Fig. 8J). FKBP52 has PPIase activity that mediates structural alterations, which act as molecular switches. It elicits diverse cellular functions, including protein folding, cellular signaling, apoptosis, and transcription. Our results show that the PPIase activity of FKBP52 is also required for the increased stability of ERα, suggesting that alterations in higher-order structures of ERα as a result of FKBP52-mediated prolyl isomerization might control association with BRCA1, affecting protein stability of ERα.
In the present study, we also showed that FKBP51 and FKBP52 have opposing effects on ERα stability and function. This is consistent with findings of previous studies showing that FKBP51 functions in the opposite way to FKBP52 for GR and PR (55, 56). In addition, results of association studies suggest that high expression of FKBP5 is related to better prognosis of breast cancer, which is also opposite to poor prognosis of breast cancer with high expression of FKBP4. We speculate that FKBP51 might compete with FKBP52 to bind to ERα and destabilize its expression. We also investigated the expression levels of FKBP4 and FKBP5 in various cancers other than breast cancer and found that FKBP4 is overexpressed in most types of cancer, whereas FKBP5 is more abundantly expressed in normal tissues than in cancer. These results suggest that FKBP51 and FKBP52 may have many common targets other than ERα but function in the opposite way to strictly control their functions and regulate proliferation. In contrast, both FKBP51 and FKBP52 were shown to positively regulate the activity of AR (28, 55), suggesting that these two FKBPs have some overlapping functions. Consistent with the latter notion, mice deficient in FKBP51 or FKBP52 alone are viable (57), whereas those deficient in both proteins manifest embryonic mortality (56). The regulation of target factors by FKBP51 and FKBP52 is complex. Given that ERα is a major therapeutic target for breast cancer, a more detailed analysis is required to understand how FKBP51 and FKBP52 contribute to ERα stabilization.
Materials and Methods
Candidate Isolation.
Protein interaction data were obtained using the IntAct (58) and BioGRID (59). We considered the protein listed in at least one database as an interactor. The Pearson correlation coefficient was calculated using mRNA expression data from TCGA BRCA. Fold-changes in mRNA expression between normal tissue and ERα-positive breast cancer were calculated by means of these groups. HRs were obtained from the Kaplan-Meier Plotter. In the case of genes corresponding to several probes, we considered as candidates those with at least one probe meeting our criteria.
Kaplan-Meier Survival Analysis.
Kaplan-Meier relapse-free survival plots were generated using the 2020 version of the Kaplan-Meier Plotter for breast cancer (60). Patients were split by median mRNA expression. Affymetrix probes 200895_s_at and 224856_at were used as probes for FKBP52 and FKBP51, respectively. ER status included “derive ER status from gene expression data” (60). All datasets available in 2020 were used for the analysis. For a comprehensive analysis, we analyzed 19,462 probes (12,180 genes) from the largest datasets (i.e., E-MTAB-365, GSE11121, GSE12093, GSE12276, GSE1456, GSE16391, GSE16446, GSE16716, GSE17705, GSE17907, GSE19615, GSE20271, GSE2034, GSE20685, GSE20711, GSE21653, GSE2603, GSE26971, GSE2990, GSE31519, GSE3494, GSE37946, GSE42568, GSE45255, GSE4611, GSE4922, GSE5327, GSE6532, GSE7390, and GSE9195). Significance was calculated using a two-tailed log-rank test with P < 0.05 considered significant.
Immunoblotting.
To prepare the total cell lysates, the collected cells were washed with ice-cold phosphate-buffered saline (PBS), suspended in sample buffer (2% sodium dodecyl sulfate [SDS], 10% glycerol, 100 μM dithiothreitol, 0.1% bromophenol blue, and 50 mM Tris⋅HCl at pH 6.8), and boiled for 5 min. Raw digital images were captured using a ChemiDoc Imaging Systems (Bio-Rad). The bands of the target protein were quantified using Image Lab (Bio-Rad) and normalized to that of β-actin, unless stated otherwise. Representative images are presented in the figures. The decay curve of ERα was plotted using GraphPad Prism version 9 (GraphPad Software) based on the band intensity of β-actin. The exponential one-phase decay equation from the nonlinear regression was used to generate the decay curve.
Immunoprecipitation.
Immunoprecipitation was performed as previously described (61). Cells were lysed in immunoprecipitation kinase buffer (50 mM Hepes–NaOH at pH 8.0, 150 mM NaCl, 2.5 mM ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid, 1 mM dithiothreitol, 0.1% Tween-20, and 10% glycerol) or radioimmunoprecipitation assay buffer (50 mM Tris⋅HCl at pH 7.5, 150 mM NaCl, 1 mM ethylenediaminetetraacetic acid, 1% Triton X-100, 10% sodium deoxycholate, and 10% SDS) supplemented with protease inhibitors (phenylmethylsulfonyl fluoride, leupeptin, pepstatin A, and aprotinin), and phosphatase inhibitors (50 mM NaF, 0.1 mM Na3VO4, 15 mM p-nitrophenylphosphate, and 80 mM β-glycerophosphate). The lysates were incubated with FLAG-M2 agarose (A2220, Sigma-Aldrich), or immunoprecipitation was performed with various antibodies for 1 h at 4 °C with rotation.
Antibodies.
The antibodies used in this study were BRCA1 (sc6954, Santacruz), ERα (cs8644, Cell Signaling Technology), FKBP51 (ab126715, Abcam), FKBP52 (10655-1-AP, Abcam), FLAG (M185-3L, MBL), β-actin (ab6276, Abcam), Hsp90 (sc13119, Santa Cruz Biotechnology), and H2B (ab1790, Abcam).
RT-qPCR.
Total RNA was extracted as previously described (62). Briefly, total RNA was extracted using ISOGEN II (311-07361, Nippon Gene) according to the manufacturer’s protocol, and reverse transcription was performed. RNA was reverse transcribed with random primers using the High-Capacity cDNA Reverse Transcription Kit (4368814, ABI). qPCR was performed using FastStart Universal SYBR Green Master Mix (11226200, Roche) and a StepOnePlus real-time PCR system (Applied Biosystems). Expression levels were normalized to that of glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The following primers were used for amplification: ESR1-F, TGATGAAAGGTGGGATACGA; ESR1-R, AAGGTTGGCAGCTCTCATGT; GAPDH-F, GAGTCAACGGATTTGGTCGT; and GAPDH-R, TTGATTTTGGAGGGATCTCG. All sequences are shown in the 5′ to 3′ direction.
Lentivirus Generation and Infection.
Lentivirus generation and infection were performed as described previously (63). Briefly, lentiviruses expressing FKBP52, FKBP51, shControl, shFKBP52, shFKBP51, shBRCA1, and shERα were generated by cotransfection of HEK293T cells with psPAX2 and pMD2.G, and the respective CSII-CMV-MCS-IRES2-Bsd or CS-RfA-ETBsd, using Polyethylenimine (PEI) MAX (24765-1, Polysciences). Cells infected with viruses were treated with 10 μg/mL blasticidin (A1113903, Gibco) for 2 d. To drive expression of shRNA, doxycycline (Dox; D9891, Sigma-Aldrich) was added to the medium at a concentration of 1 μg/mL
RNA-Sequencing Analysis.
MCF7 cells expressing shControl, shFKBP52, or shFKBP51 were cultured in the presence of Dox for 2 d and then subjected to total RNA extraction. RNA-seq analysis was performed as previously described (61).
Quantification and Statistical Analysis.
Quantitative data are presented as mean ± SEM. Two-group comparisons were performed with t test. To compare three or more groups, one-way ANOVA followed by Dunnett multiple comparison test was performed for multiple comparisons. Results were considered statistically significant at P < 0.05, P < 0.01, P < 0.001, and P < 0.0001. Statistical analyses were performed using GraphPad Prism, version 9.
Mouse Xenograft.
Protocols were prepared in accordance with the regulations on animal use at Yamaguchi University and were approved by the Committee for Animal Use of Yamaguchi University. Female NOD/Shi-scid IL-2RγKO mice (In-Vivo Science Inc.) were maintained under specific pathogen-free conditions and fed a sterilized standard diet containing 600 mg/kg Dox (CE-2, CLEA Japan Inc.). Subcutaneous implantation of 17β-estradiol pellets (1.7 mg, 90-d release; NE-121, Innovative Research of America) and ovariectomy were performed at 7 wk of age. One week postoperation, MCF7 cells expressing shControl or shFKBP52 (5 × 106 cells/100 μL PBS per site) were subcutaneously transplanted into the left or right side of the flank, respectively. The tumor sizes were measured twice weekly using a digital caliper. If a tumor was undetected, the tumor size was considered to be 0. The tumor volumes were estimated using the following formula: (short diameter)2 × (longest diameter) × 0.5.
Immunohistochemistry.
Engrafted tumor tissues were fixed in 10% phosphate-buffered formalin and embedded in paraffin. Sections 2 μm thick were obtained for immunohistochemistry. Sections were deparaffinized and subjected to antigen retrieval by boiling in pH 9 antigen retrieval solution (415291, Nichirei Bioscience) for 20 min in a pressure cooker. Endogenous peroxidase activity was inhibited by soaking the samples in 3% hydrogen peroxide (081-04215, Wako). Blocking was performed using a mixture of 2% bovine serum albumin and 10% skim milk for 30 min at room temperature (20–25 °C). Next, sections were incubated with a rat anti–Ki-67 antibody (dilution 1:1,000; 14–5698-82, Thermo Fisher Scientific) for 60 min at room temperature. After incubating with anti-rat immunoglobulins conjugated to peroxidase-labeled micropolymer (ImmPRESS HRP Reagent Kit, Vector Laboratories Inc.) for 30 min at room temperature, the immunoreaction was visualized using a DAB substrate kit (11718096001, Roche). Finally, the sections were counterstained with Mayer’s hematoxylin. Whole-slide images were obtained using a NanoZoomer 2.0RS (Hamamatsu Photonics). The percentage of Ki-67–positive cells was determined using the function of “Positive cell detection” in QuPath, version 0.3.1, software (64). Other detailed information is described in SI Appendix.
Supplementary Material
Acknowledgments
We thank N. Kawasaki and K. Maeda for technical assistance; Dr. KI Nakayama (Kyusyu University) for critical discussions; and Drs. M. Okada (Tokyo University of Technology), T. Ohta (St. Marianna University School of Medicine), M. Iizuka (Teikyo University), N. Saitoh (The Cancer Institute of Japanese Foundation for Cancer Research), T. Fujita (The Cancer Institute of Japanese Foundation for Cancer Research), and Y. Johmura (Tokyo University) for materials and/or discussions. We also thank the Yamaguchi University Project for the formation of the Core Research Center. This study was supported in part by Grants-in-Aid for Scientific Research (KAKENHI) Grants 18H02681 and 20K21503 from the Ministry of Education, Culture, Sports, Science, and Technology of Japan Fusion Oriented Research for Disruptive Science and Technology; a grant from the MSD Life Science Foundation; and a grant from the Shinnihon Foundation of Advanced Medical Treatment Research.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2110256119/-/DCSupplemental.
Data Availability
RNA-sequencing raw reads have been submitted to the DNA Data Bank of Japan Sequence Read Archive, the National Center for Biotechnology Information Sequence Read Archive, and the European Bioinformatics Institute Sequence Read Archive databases under accession number DRA011728 (65).
References
- 1.Fuentes N., Silveyra P., Estrogen receptor signaling mechanisms. Adv. Protein Chem. Struct. Biol. 116, 135–170 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cook K. L., Shajahan A. N., Clarke R., Autophagy and endocrine resistance in breast cancer. Expert Rev. Anticancer Ther. 11, 1283–1294 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saha Roy S., Vadlamudi R. K., Role of estrogen receptor signaling in breast cancer metastasis. Int. J. Breast Cancer 2012, 654698 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Musgrove E. A., Sutherland R. L., Biological determinants of endocrine resistance in breast cancer. Nat. Rev. Cancer 9, 631–643 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Schiff R., Massarweh S., Shou J., Osborne C. K., How growth factor signaling and estrogen receptor coregulators modulate response Clin. Cancer. Res. 9, 447s–454s (2003). [PubMed] [Google Scholar]
- 6.Tecalco-Cruz A. C., Ramírez-Jarquín J. O., Cruz-Ramos E., Estrogen receptor alpha and its ubiquitination in breast cancer cells. Curr. Drug Targets 20, 690–704 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Dhamad A. E., Zhou Z., Zhou J., Du Y., Systematic proteomic identification of the heat shock proteins (Hsp) that interact with estrogen receptor alpha (ERα) and biochemical characterization of the ERα-Hsp70 interaction. PLoS One 11, e0160312 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tecalco-Cruz A. C., Pérez-Alvarado I. A., Ramírez-Jarquín J. O., Rocha-Zavaleta L., Nucleo-cytoplasmic transport of estrogen receptor alpha in breast cancer cells. Cell. Signal. 34, 121–132 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Hewitt S. C., Korach K. S., Estrogen receptors: New directions in the new millennium. Endocr. Rev. 39, 664–675 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun J., Zhou W., Kaliappan K., Nawaz Z., Slingerland J. M., ERα phosphorylation at Y537 by Src triggers E6-AP-ERα binding, ERα ubiquitylation, promoter occupancy, and target gene expression. Mol. Endocrinol. 26, 1567–1577 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan M., Park A., Nephew K. P., CHIP (carboxyl terminus of Hsc70-interacting protein) promotes basal and geldanamycin-induced degradation of estrogen receptor-alpha. Mol. Endocrinol. 19, 2901–2914 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Bhatt S., Xiao Z., Meng Z., Katzenellenbogen B. S., Phosphorylation by p38 mitogen-activated protein kinase promotes estrogen receptor α turnover and functional activity via the SCF(Skp2) proteasomal complex. Mol. Cell. Biol. 32, 1928–1943 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saji S., et al. , MDM2 enhances the function of estrogen receptor alpha in human breast cancer cells. Biochem. Biophys. Res. Commun. 281, 259–265 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Wang S., et al. , RNF8 identified as a co-activator of estrogen receptor α promotes cell growth in breast cancer. Biochim. Biophys. Acta Mol. Basis Dis. 1863, 1615–1628 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Zhu J., et al. , The atypical ubiquitin ligase RNF31 stabilizes estrogen receptor α and modulates estrogen-stimulated breast cancer cell proliferation. Oncogene 33, 4340–4351 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhuang T., et al. , SHARPIN stabilizes estrogen receptor α and promotes breast cancer cell proliferation. Oncotarget 8, 77137–77151 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eakin C. M., Maccoss M. J., Finney G. L., Klevit R. E., Estrogen receptor alpha is a putative substrate for the BRCA1 ubiquitin ligase. Proc. Natl. Acad. Sci. U.S.A. 104, 5794–5799 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harding M. W., Galat A., Uehling D. E., Schreiber S. L., A receptor for the immunosuppressant FK506 is a cis-trans peptidyl-prolyl isomerase. Nature 341, 758–760 (1989). [DOI] [PubMed] [Google Scholar]
- 19.Siekierka J. J., Hung S. H., Poe M., Lin C. S., Sigal N. H., A cytosolic binding protein for the immunosuppressant FK506 has peptidyl-prolyl isomerase activity but is distinct from cyclophilin. Nature 341, 755–757 (1989). [DOI] [PubMed] [Google Scholar]
- 20.Bierer B. E., et al. , Two distinct signal transmission pathways in T lymphocytes are inhibited by complexes formed between an immunophilin and either FK506 or rapamycin. Proc. Natl. Acad. Sci. U.S.A. 87, 9231–9235 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu K. P., Finn G., Lee T. H., Nicholson L. K., Prolyl cis-trans isomerization as a molecular timer. Nat. Chem. Biol. 3, 619–629 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Dolinski K., et al. , Functions of FKBP12 and mitochondrial cyclophilin active site residues in vitro and in vivo in Saccharomyces cerevisiae. Mol. Biol. Cell 8, 2267–2280 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tong M., Jiang Y., FK506-binding proteins and their diverse functions. Curr. Mol. Pharmacol. 9, 48–65 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riggs D. L., et al. , The Hsp90-binding peptidylprolyl isomerase FKBP52 potentiates glucocorticoid signaling in vivo. EMBO J. 22, 1158–1167 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheung-Flynn J., et al. , Physiological role for the cochaperone FKBP52 in androgen receptor signaling. Mol. Endocrinol. 19, 1654–1666 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Tranguch S., et al. , Cochaperone immunophilin FKBP52 is critical to uterine receptivity for embryo implantation. Proc. Natl. Acad. Sci. U.S.A. 102, 14326–14331 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Z., et al. , FK506-binding protein 52 is essential to uterine reproductive physiology controlled by the progesterone receptor A isoform. Mol. Endocrinol. 20, 2682–2694 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maeda K., et al. , FKBP51 and FKBP52 regulate androgen receptor dimerization and proliferation in prostate cancer cells. Mol. Oncol. 16, 940–956 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Denny W. B., Valentine D. L., Reynolds P. D., Smith D. F., Scammell J. G., Squirrel monkey immunophilin FKBP51 is a potent inhibitor of glucocorticoid receptor binding. Endocrinology 141, 4107–4113 (2000). [DOI] [PubMed] [Google Scholar]
- 30.Hubler T. R., et al. , The FK506-binding immunophilin FKBP51 is transcriptionally regulated by progestin and attenuates progestin responsiveness. Endocrinology 144, 2380–2387 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Ni L., et al. , FKBP51 promotes assembly of the Hsp90 chaperone complex and regulates androgen receptor signaling in prostate cancer cells. Mol. Cell. Biol. 30, 1243–1253 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Febbo P. G., et al. , Androgen mediated regulation and functional implications of fkbp51 expression in prostate cancer. J. Urol. 173, 1772–1777 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Federer-Gsponer J. R., et al. , Delineation of human prostate cancer evolution identifies chromothripsis as a polyclonal event and FKBP4 as a potential driver of castration resistance. J. Pathol. 245, 74–84 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Li L., Lou Z., Wang L., The role of FKBP5 in cancer aetiology and chemoresistance. Br. J. Cancer 104, 19–23 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hong C., et al. , Elevated FKBP52 expression indicates a poor outcome in patients with breast cancer. Oncol. Lett. 14, 5379–5385 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berry N. B., Fan M., Nephew K. P., Estrogen receptor-alpha hinge-region lysines 302 and 303 regulate receptor degradation by the proteasome. Mol. Endocrinol. 22, 1535–1551 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sinars C. R., et al. , Structure of the large FK506-binding protein FKBP51, an Hsp90-binding protein and a component of steroid receptor complexes. Proc. Natl. Acad. Sci. U.S.A. 100, 868–873 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schülke J. P., et al. , Differential impact of tetratricopeptide repeat proteins on the steroid hormone receptors. PLoS One 5, e11717 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsuboi K., et al. , Different epigenetic mechanisms of ERα implicated in the fate of fulvestrant-resistant breast cancer. J. Steroid Biochem. Mol. Biol. 167, 115–125 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Á. Bartha, B. Győrffy, TNMplot.com: A web tool for the comparison of gene expression in normal, tumor and metastatic tissues. bioRxiv [Preprint] (2020). 10.1101/2020.11.10.376228 (Accessed 5 February 2021). [DOI] [PMC free article] [PubMed]
- 41.Medvar B., Raghuram V., Pisitkun T., Sarkar A., Knepper M. A., Comprehensive database of human E3 ubiquitin ligases: Application to aquaporin-2 regulation. Physiol. Genomics 48, 502–512 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fu M., Wang C., Li Z., Sakamaki T., Pestell R. G., Minireview: Cyclin D1: Normal and abnormal functions. Endocrinology 145, 5439–5447 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Qie S., Diehl J. A., Cyclin D1, cancer progression, and opportunities in cancer treatment. J. Mol. Med. (Berl.) 94, 1313–1326 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lundberg A., et al. , The long-term prognostic and predictive capacity of cyclin D1 gene amplification in 2305 breast tumours. Breast Cancer Res. 21, 34 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu B., Fan Y., CDK4/6 inhibition in early-stage breast cancer: How far is it from becoming standard of care? Lancet Oncol. 22, 159–160 (2021). [DOI] [PubMed] [Google Scholar]
- 46.Chen W., et al. , Grainyhead-like 2 enhances the human telomerase reverse transcriptase gene expression by inhibiting DNA methylation at the 5′-CpG island in normal human keratinocytes. J. Biol. Chem. 285, 40852–40863 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dompe N., et al. , A whole-genome RNAi screen identifies an 8q22 gene cluster that inhibits death receptor-mediated apoptosis. Proc. Natl. Acad. Sci. U.S.A. 108, E943–E951 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Werner S., et al. , Dual roles of the transcription factor grainyhead-like 2 (GRHL2) in breast cancer. J. Biol. Chem. 288, 22993–23008 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holding A. N., et al. , VULCAN integrates ChIP-seq with patient-derived co-expression networks to identify GRHL2 as a key co-regulator of ERa at enhancers in breast cancer. Genome Biol. 20, 91 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Helzer K. T., et al. , The phosphorylated estrogen receptor α (ER) cistrome identifies a subset of active enhancers enriched for Direct ER-DNA binding and the transcription factor GRHL2. Mol. Cell. Biol. 39, e00417-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zajchowski D. A., Sager R., Webster L., Estrogen inhibits the growth of estrogen receptor-negative, but not estrogen receptor-positive, human mammary epithelial cells expressing a recombinant estrogen receptor. Cancer Res. 53, 5004–5011 (1993). [PubMed] [Google Scholar]
- 52.Gajulapalli V. N. R., Malisetty V. L., Chitta S. K., Manavathi B., Oestrogen receptor negativity in breast cancer: A cause or consequence? Biosci. Rep. 36, e00432 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ma Y., et al. , BRCA1 regulates acetylation and ubiquitination of estrogen receptor-alpha. Mol. Endocrinol. 24, 76–90 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hosey A. M., et al. , Molecular basis for estrogen receptor alpha deficiency in BRCA1-linked breast cancer. J. Natl. Cancer Inst. 99, 1683–1694 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ratajczak T., Cluning C., Ward B. K., Steroid receptor-associated immunophilins: A gateway to steroid signalling. Clin. Biochem. Rev. 36, 31–52 (2015). [PMC free article] [PubMed] [Google Scholar]
- 56.Storer C. L., Dickey C. A., Galigniana M. D., Rein T., Cox M. B., FKBP51 and FKBP52 in signaling and disease. Trends Endocrinol. Metab. 22, 481–490 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yong W., et al. , Essential role for co-chaperone Fkbp52 but not Fkbp51 in androgen receptor-mediated signaling and physiology. J. Biol. Chem. 282, 5026–5036 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Orchard S., et al. , The MIntAct project—IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 42, D358–D363 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oughtred R., et al. , The BioGRID database: A comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci. 30, 187–200 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Györffy B., et al. , An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat. 123, 725–731 (2010). [DOI] [PubMed] [Google Scholar]
- 61.Masaki T., et al. , Calcineurin regulates the stability and activity of estrogen receptor α. Proc. Natl. Acad. Sci. U.S.A. 118, e2114258118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shimada M., et al. , Chk1 is a histone H3 threonine 11 kinase that regulates DNA damage-induced transcriptional repression. Cell 132, 221–232 (2008). [DOI] [PubMed] [Google Scholar]
- 63.Goshima T., et al. , Calcineurin regulates cyclin D1 stability through dephosphorylation at T286. Sci. Rep. 9, 12779 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bankhead P., et al. , QuPath: Open source software for digital pathology image analysis. Sci. Rep. 7, 16878 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.M. Habara, M. Shimada, Transcriptome data of MCF7 cells expressing doxycycline-inducible FKBP51, FKBP52, and control shRNAs. DNA Data Bank of Japan. https://ddbj.nig.ac.jp/resource/sra-submission/DRA011728. Deposited 14 March 2021. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-sequencing raw reads have been submitted to the DNA Data Bank of Japan Sequence Read Archive, the National Center for Biotechnology Information Sequence Read Archive, and the European Bioinformatics Institute Sequence Read Archive databases under accession number DRA011728 (65).