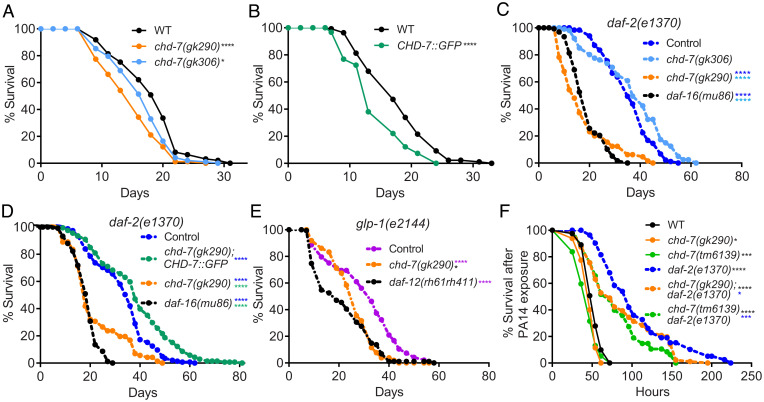
Fig. 2.
chd-7 affects longevity and response to pathogen. (A–E) chd-7 promotes longevity in wild-type, daf-2(e1370), and glp-1(e2144) mutants. Mean survival days on OP50-1; survival data analyzed using Kaplan−Meier test. For all experiments, the asterisk in color represents strain of reference for statistical analysis. Details of number of animals and additional data from replicates can be found in SI Appendix, Table S1. (A) WT (18.12), chd-7(gk290) (14.91), chd-7(gk306) (16.53). *P < 0.05 and ****P < 0.0001 compared to the wild-type, N2 strain. (B) WT (17.85), CHD-7::GFP (14.35). ****P < 0.0001 compared to the wild-type, N2 strain. (C) daf-2(e1370) (35.58), chd-7(gk306);daf-2(e1370) (37.18), chd-7(gk290);daf-2(e1370) (17.47), daf-16(mu86);daf-2(e1370) (18.85). ****P < 0.0001. (D) daf-2(e1370) (27.9), chd-7(gk290);daf-2(e1370);CHD-7::GFP (30.84), chd-7(gk290);daf-2(e1370) (19.13), daf-16(mu86);daf-2(e1370) (14.48). ****P < 0.0001. (E) glp-1(e2144) (30.4), chd-7(gk290);glp-1(e2144) (28.37), glp-1(e2144);daf-12(rh61rh411) (25.99). *P < 0.05 and ****P < 0.0001. (F) chd-7 mediates the response against the opportunistic bacteria P. aeruginosa. Mean lifespan in hours (m) ± SEM. n is the number of animals analyzed/total number in experiment. WT (mean = 52.11 ± 0.96, n = 94 of 162), chd-7(gk290) (mean = 47.45 ± 1.05, n = 54 of 130), chd-7(tm6139) (mean = 44.47 ± 1.57, n = 56 of 80), daf-2(e1370) (mean = 105.46 ± 5.61, n = 65 of 115), chd-7(gk290);daf-2(e1370) (mean = 86.74 ± 4.21, n = 103 of 120), and chd-7(tm6139);daf-2(e1370) (mean = 79.13 ± 4.9, n = 50 of 60). *P < 0.05; ***P < 0.001 and ****P < 0.0001. Survival data analyzed using Kaplan−Meier test.