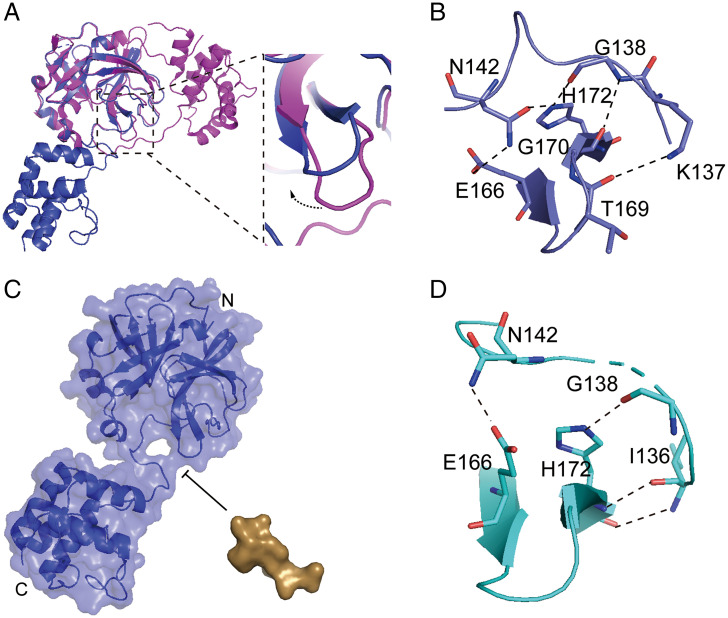
Fig. 4.
The conformation of monomeric Mpro is not suitable for the substrate binding. (A) Superposition of the extended monomeric structure and one protomer of dimeric structure based on the N-terminal domain, with the extended structure colored in blue, and dimer structure colored in magenta (Left) and close-up view of the conformational change of the β-turn (Right). (B) The interaction between the β-turn (E166-H172) and the active loop (K137-N142) of the extended monomeric structure (blue). (C) The model of the extended monomer Mpro and substrate (TSAVLQ, derived from the N-terminal autocleavage sequence of the viral protease), the volume of substrate bound pocket is insufficient to accommodate the substrate. The extended monomer Mpro and substrate are shown as surface, with the monomeric Mpro colored in blue and substrate colored in brown. (D) The interaction between the β-turn (E166-H172) and the active loop (K136-N142) of the compact monomeric structure (cyan). The key residues involved in interaction are shown as stick models. Polar interactions are indicated with black dashed lines.