Fig. 1.
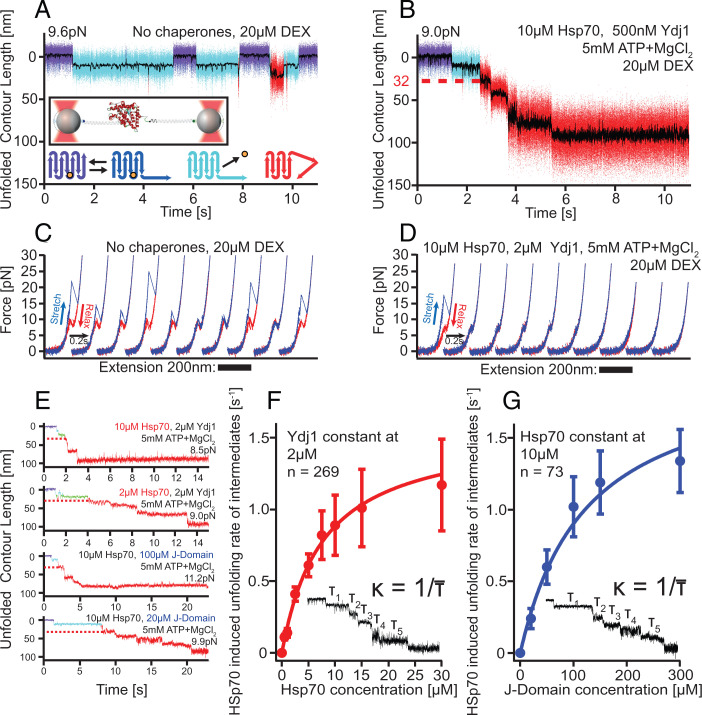
Hsp70/40 unfolds GR-LBD in a stepwise manner. (A) In the absence of chaperones, holo GR-LBD shows fast opening and closing of the N-terminal “lid” (fast transitions between purple and dark-blue state), ligand dissociation (transition to light-blue state), and ligand rebinding (return to purple/dark-blue flipping) as well as rare partial unfoldings (red) (22). Inset: Experimental scheme for the single-molecule optical tweezers experiment. Bottom: Graphical illustration of states in the upper trace (same color code). The orange sphere symbolizes DEX. (B) Sample trace of Hsp70/40 unfolding apo GR-LBD completely via five intermediates within ∼5 s. Unfolding sets in within ∼1 s after DEX dissociation. The red dashed line marks the 32 nm of unfolded contour length at which the first chaperone-induced unfolding intermediate is located. (C) Stretch-relax cycles of the GR-LBD in the absence of chaperones showing the unfolding and refolding fingerprint of GR-LBD (22). Ten consecutive pulls of the same molecule are presented, including unfolding from ligand-bound states (cycles 1, 4, 8, and 10) and ligand-unbound states (cycles 2, 3, 5, 6, 7, and 9). (D) Ten consecutive traces starting from a natively folded GR-LBD in the presence of Hsp70/40. (E) Sample traces of unfolding at various chaperone concentrations. In all traces, the first 32-nm intermediate is visible (red dashed line). (F) Unfolding rates (inverse of average dwell time per step) versus Hsp70 concentration at constant Ydj1 concentration. Error bars show SEM. (G) Unfolding rates (inverse of average dwell time per step) versus J-domain concentration at constant Hsp70 concentration. Error bars show SEM.