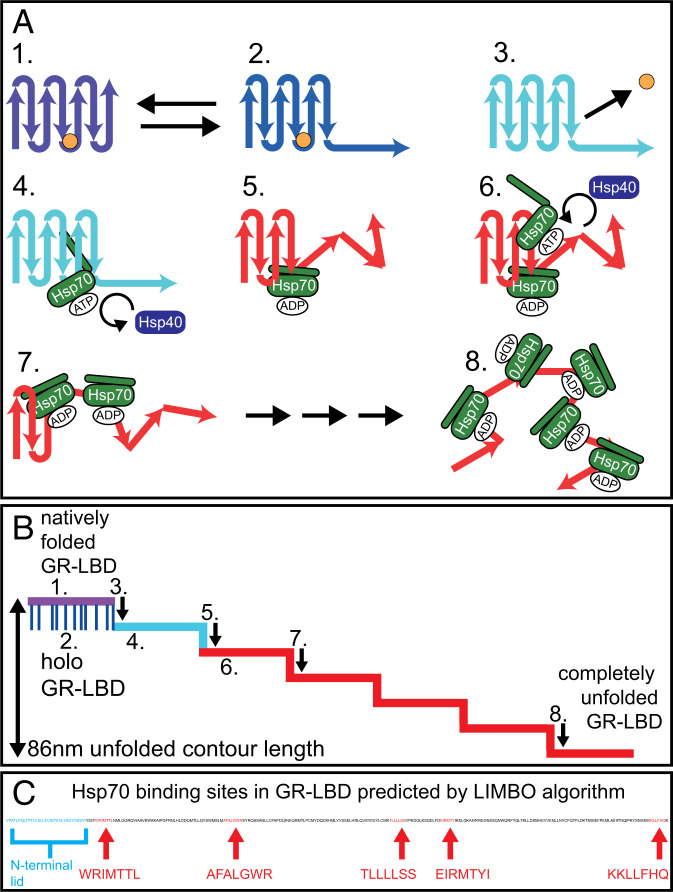
Fig. 4.
Model of Hsp70/40-induced unfolding of GR-LBD. The same color code as in all previous passive-mode traces was used to identify the states of GR-LBD: Purple: natively folded, DEX bound, lid closed; dark-blue: DEX bound, lid open; light-blue: DEX unbound, apo structure still folded; red: unfolded by chaperones. (A, 1) GR-LBD is in the natively folded holo state with its N-terminal lid closed (purple state). The hormone DEX is depicted as a small orange circle. (A, 2) Holo GR-LBD undergoes rapid flipping transitions between the lid-closed (purple) and the lid-open (dark-blue) states. (A, 3) After DEX dissociation, the lid (i.e., the first 33 aa) remains unfolded permanently (light-blue state). (A, 4) Chaperones can now attack apo GR-LBD. The first Hsp70 (in its open ATP bound state) binds to the 32-nm binding motif, which is located in the folded remainder of apo GR-LBD. 5.) Upon Hsp40-stimulated ATP hydrolysis, Hsp70 unfolds (“chews open”) the N-terminal upstream part of apo GR-LBD until the 32-nm binding motif. The associated step in contour length occurs almost simultaneously with ATP hydrolysis. Hsp70 then remains bound to the unfolded peptide chain in the ADP-bound state until ADP dissociation and rebinding of a new ATP occurs (minutes). (A, 6–8) Consecutive binding of up to four more Hsp70 molecules and J-protein–catalyzed ATP hydrolysis induced further unfoldings. The mechanism may also involve a combination of ratchet and/or entropic pulling mechanisms. At the end of the unfolding process, GR-LBD is a completely unfolded peptide chain decorated with up to five Hsp70s bound in the ADP state. The unfolding most likely starts at the N terminus, but does not necessarily have to proceed sequentially as depicted. (B) Schematic illustration of a typical Hsp70/40-induced unfolding of GR-LBD. Steps 1–8 are mapped according to the model in A. (C) Position and sequence of the Hsp70-binding sites in the sequence of GR-LBD as predicted by the LIMBO algorithm (27).