Fig. 3.
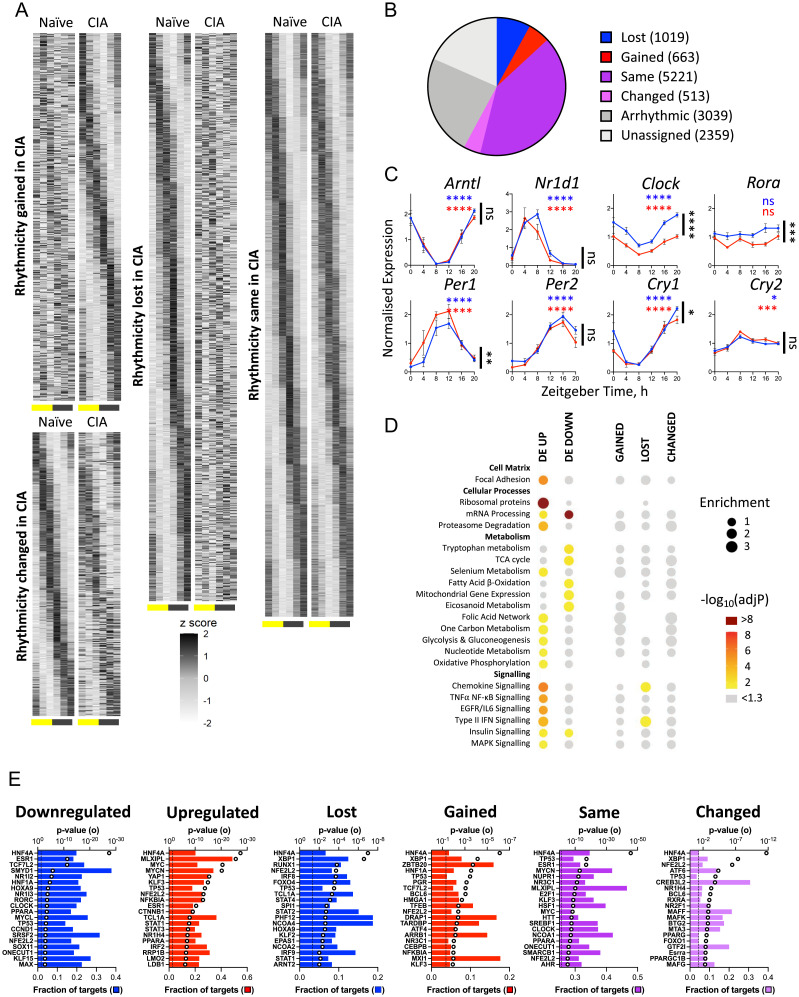
The liver transcriptome shows circadian perturbation in response to arthritis. (A and B) Matched liver samples were analyzed by RNA sequencing to characterize the effect of distal inflammatory disease. The majority of genes were rhythmic under at least one condition, and genes were grouped according to change or maintenance of rhythmicity with CIA using compareRhythms (Fig. 2). (C) Rhythmic expression of most core clock genes was maintained in liver, although expression of Clock and Rora was significantly reduced with disease (CIA vs. naïve comparison by two-way ANOVA in black; adjusted P [adjP] value of JTK analysis in blue and red; n = 5/point. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Data are presented as mean ± SEM). (D) Groups of genes showing significant differential expression (DE) or rhythmic change with disease were analyzed using the Enrichr tool (Materials and Methods and Fig. 2). Enrichment of metabolic and signaling categories were predominantly associated with changes in expression level rather than rhythmicity. (E) Upstream regulator analysis of liver transcripts differentially expressed with CIA identified metabolic and inflammatory mediators. The top 20 most significant regulators are shown for each category. Open circle represents significance of enrichment; filled bar represents fraction of downstream targets found in the category; dashed line indicates P = 0.05. ns, not significant.