Extended Data Fig. 1. Strain engineering and 4EPS quantification.
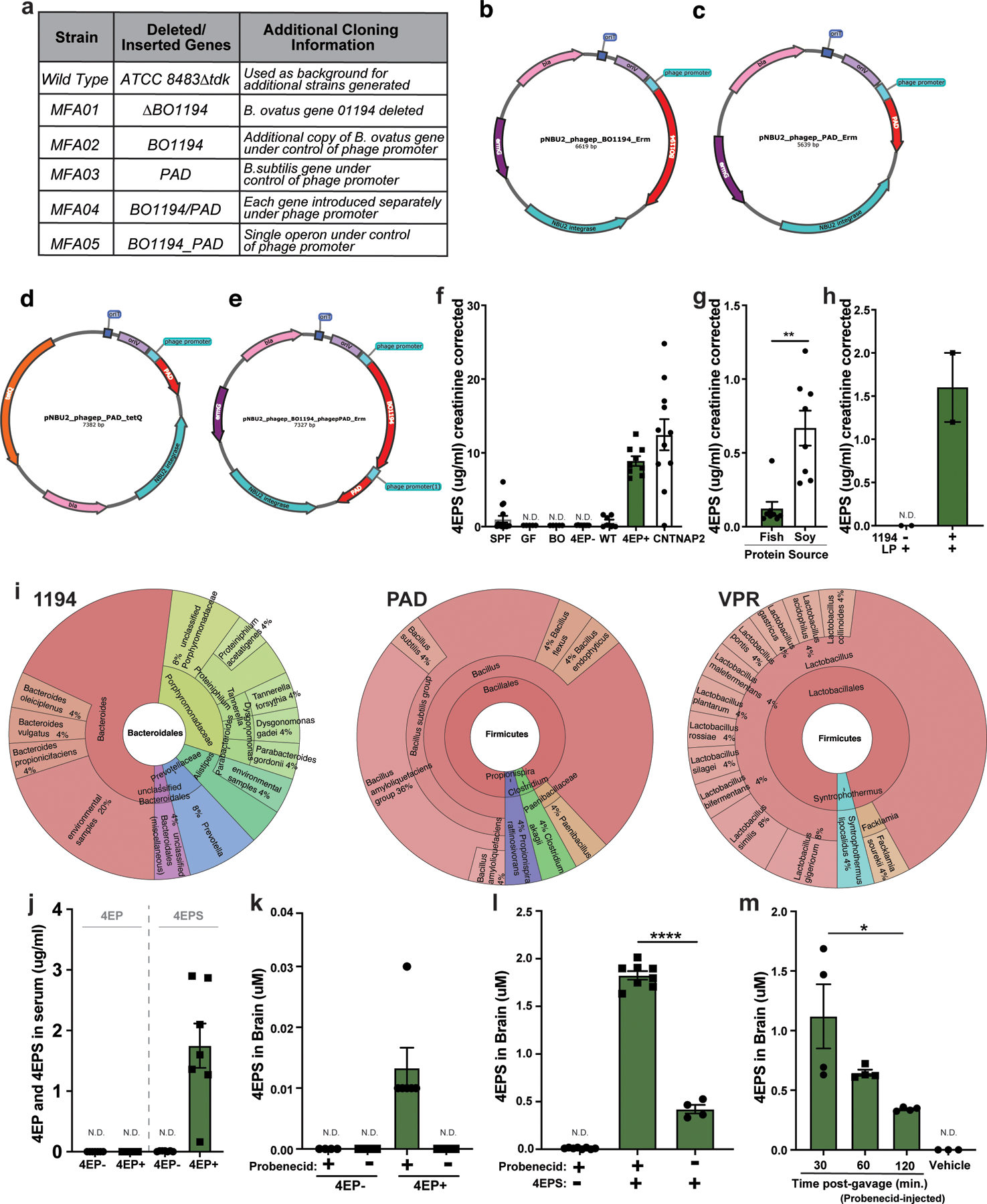
a, Strain legend corresponding to Figure 1b-d, containing further details for strain names and cloning strategy. Additional information can be found in Methods and Supplementary Information Tables 1-3. b-e, Maps for integration vectors. b, The BO1194 gene was cloned into the pNBU2 vector with an erythromycin resistant marker under the phage promoter. c, The PAD gene was cloned into pNBU2 vector with an erythromycin resistant marker under the phage promoter. d, The PAD gene was cloned into the pNBU2 vector with a tetracycline resistant marker under the phage promoter. e, BO1194 and PAD were tandemly connected, putting the phage promoter in front of both genes, then icloned into the pNBU2 vector with the erythromycin resistant marker. f, Creatinine corrected urinary levels of 4EPS from SPF, GF, gnotobiotic mice colonized with strains as labeled, and a mouse model of atypical behavior including anxiety-like phenotypes. This data corresponds to abbreviated data in Fig. 1f. SPF, specific pathogen free mice; GF, germ-free mice; BO, Bacteroides ovatus mono-colonized mice; 4EP-, GF mice colonized with non-producing strain pair (B. ovatus Δ1194 and WT L. plantarum with endogenous VPR) used throughout; WT, GF mice colonized with wild type strain pair (WT B. ovatus and WT L. plantarum with endogenous VPR) before optimal engineering; 4EP+, colonized with strain pair engineered for higher production of 4EP+ (B. ovatus+1194/PAD and WT L. plantarum with endogenous VPR), used throughout; CNTNAP2, conventionally colonized mouse model of atypical behavior (left to right columns: n=15, 5, 5, 9, 7, 9, 11). g, 4EPS levels (creatinine corrected) in urine of wild type, specific pathogen free (SPF) mice fed one of two isocaloric diets matched for protein, mineral, carbohydrate, and fat levels but differing in either high tyrosine fish meal (fish diet), or a high plant protein (soy diet) (n=8 each group)p=0.002. h, 4EPS levels (creatinine corrected) in urine of gnotobiotic mice colonized with isogenic strain pairs of B. ovatus and wild type L. plantarum, which differ only in the presence or absence of gene 1194, and thus the tyrosine lyase activity requires to convert tyrosine to p-coumaric acid (n=2 each group). i, Alignments of genes used to engineer the 4EP synthesis pathway to the reference genomes in the WoL database, showing that these genes are found in ~25 genomes each, most of which are common human gut lineages. j, 4EP and 4EPS levels (ug/ml) in serum of colonized mice (n=7 each group). k, 4EP+ and 4EP- colonized mice were injected with the organic anion transporter to inhibit potential 4EPS transport out of the brain, then analyzed by LCMS (n=6 each group). Extended data from Figure 1G. l, Conventionally colonized (SPF) mice were injected with probenecid and then injected with high dose 4EPS (n=8) or saline vehicle (n=8), and whole brain lysate was analyzed by LCMS compared to 4EPS injection alone (n=4)p<0.0001. m, Conventionally colonized (SPF) mice were injected with probenecid and then gavaged with 4EP, then whole brains were harvested at 30-minute intervals and 4EPS levels were quantified by LCMS (n=4; vehicle group n=3)p=0.01. Two independent trials on biological replicates were used for the experiments in the figure. Abbreviations: SPF, specific pathogen-free; GF, germ-free; BO, Bacteroides ovatus; WT, wild type; LP, Lactobacillus plantarum. Data represent mean ± SEM analyzed by a two-tailed Welch’s t-test or one-way ANOVA with Dunnett multiple comparisons test as appropriate. * p ≤ 0.05; ** p ≤ 0.01; *** p ≤ 0.001; **** p ≤0.0001.
