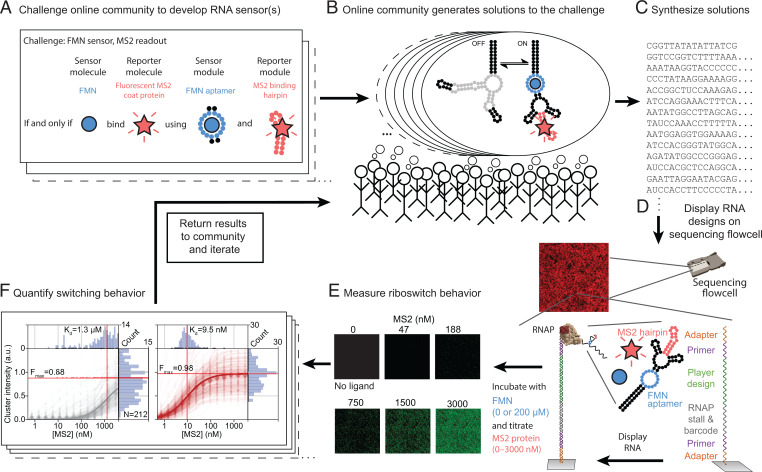
Fig. 1.
A platform for crowdsourced RNA sensor design and testing at high throughput. (A) RNA switch sensor design challenges are released to the Eterna community. In this example, an FMN aptamer (small blue circles) should be used to sense the FMN molecule (large blue circles), and in the presence of FMN, an MS2 hairpin (red circles) must fold for detection via binding of the fluorescent MS2 coat protein (red stars). (B) Eterna community designs sequences of RNA switch predicted to fold into appropriate off and on states. (C) Puzzle solutions are synthesized by DNA array synthesis and converted to libraries ready for RNA-MaP via PCR. (D) Following high-throughput sequencing of the library on an Illumina sequencing chip, a biotinylated primer is annealed and extended to create double stranded DNA. Streptavidin is then bound, and RNA polymerase (RNAP) displays one RNA switch variant solution per sequencing cluster. (E) Binding of the fluorescently labeled MS2 coat protein is then quantified across all clusters at increasing concentrations (example images are shown). Cluster sequence information is used to link fluorescent signals to underlying RNA switch sequence. (F) Binding data are quantified from multiple clusters for a single RNA switch variant (median fits are shown as the bold line, a.u. are arbitrary units). These data are then released to the Eterna community, and subsequent rounds of designs are solicited.