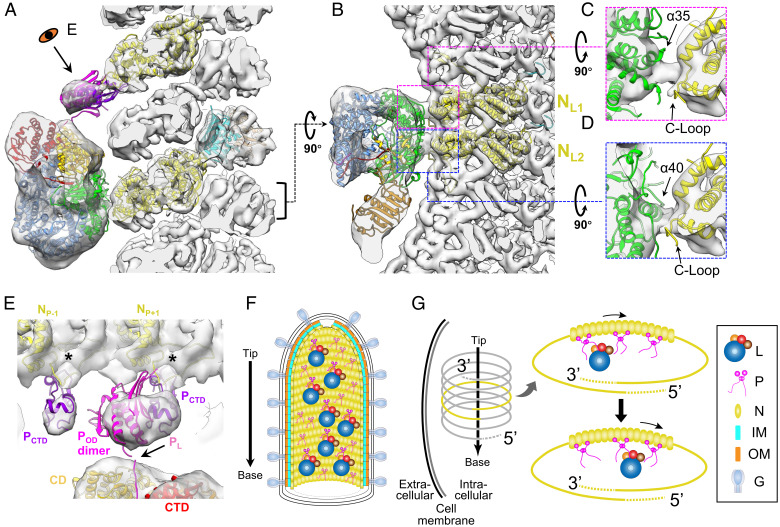
Fig. 4.
Interactions between VSV L, P, and N, and the models of multiple polymerase complexes in mature virions. (A and B) Combined 3D reconstruction (semitransparent gray) of the subnano averaged structure of VSV M-N and averaged structure of VSV polymerase complex, superimposed with atomic models (ribbon) of VSV L (PDB: 6U1X) (14), POD (PDB: 2FQM) (33), PCTD (PDB: 3HHZ) (13), N (PDB: 3HHZ, 2GIC) (12), and M (PDB: 1LG7) (31) viewed from side (A) and top (B). Following the 5′ to 3′ direction, the two N subunits involved in the interaction with L are numbered NL1, NL2 in (B). (C and D) Detailed side views of the two contacting sites between the capping domain of L and two N subunits. (E) Detailed view of interactions between P and L and between P and N. PL, POD and PCTD are shown here in pink, magenta and purple, respectively; unstructured flexible regions are not shown here. The two N subunits involved in the interaction with PCTD are numbered NP-1 and NP+1 following the 5′ to 3′ direction of the genome. (F) Schematic illustration of a mature virion with, from outer to inner, G decorated membrane, two layers of M helices, N helix, attached L and P dimers. The arrow on the left denotes the directionality of the virion. The L proteins are randomly distributed along the inside of the nucleocapsid, as are the P protein dimers. Some P dimers associate with L proteins, while the others are in unbound state. (G) A model for multiple L molecules’ movement to the 3′ end of the RNA genome by P dimers during infection. After virus entry, the nucleocapsid is released from the virion to serve as the template for subsequent transcription and replication. Multiple polymerase complexes remain attached to N. Driven by the high affinity of L to TIS at the 3′ end of the genome, L molecules are relayed by multiple P dimers all the way to the 3′ end for initiation of primary transcription.