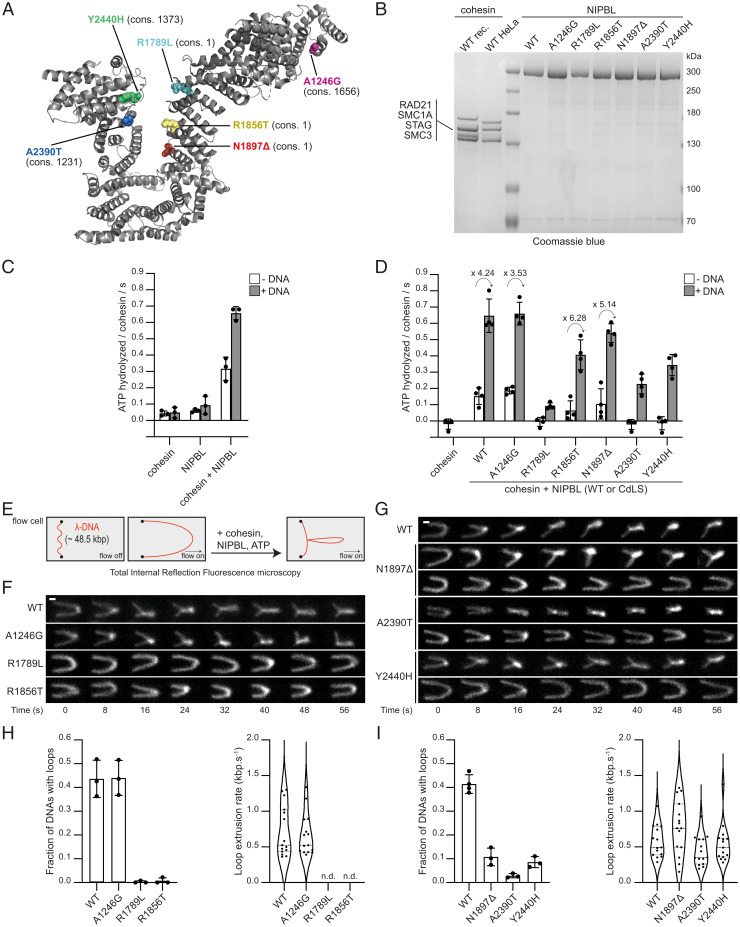
Fig. 1.
CdLS mutations in NIPBL can impair cohesin’s ATPase and DNA loop extrusion activities. (A) Structure of NIPBL (Protein Data Bank ID: 6wg3.E). Residues 1193 to 2628 are visible. Mutated residues are indicated as spheres. The evolutionary conservation ranks (cons.) among vertebrate orthologs are indicated in brackets: range 1 to 2804; 1, most conserved; 2804, least conserved. (B) Coomassie staining of cohesin and NIPBL after sodium dodecyl sulfate polyacrylamide gel electrophoresis. Subunits of recombinant (rec.) and HeLa cohesin are indicated. (C and D) Cohesin ATPase rates (mean ± SD of three and four independent experiments, for C and D, respectively) in the presence of the indicated components. The fold stimulation of ATPase activities by DNA is indicated above the arrows. (E) Cartoon illustration of the loop extrusion assay. (F and G) Stills from time-lapse recordings of representative DNA molecules in the presence of cohesin, ATP, and wild-type (WT) or mutant NIPBL. DNA was stained by Sytox Orange. (Scale bar: 1 μm.) (H and I) Frequencies of DNAs with extruded loops (Left; mean ± SD of three independent experiments) and rates of loop extrusion (Right; n.d.: not determined; median and quartiles are shown).