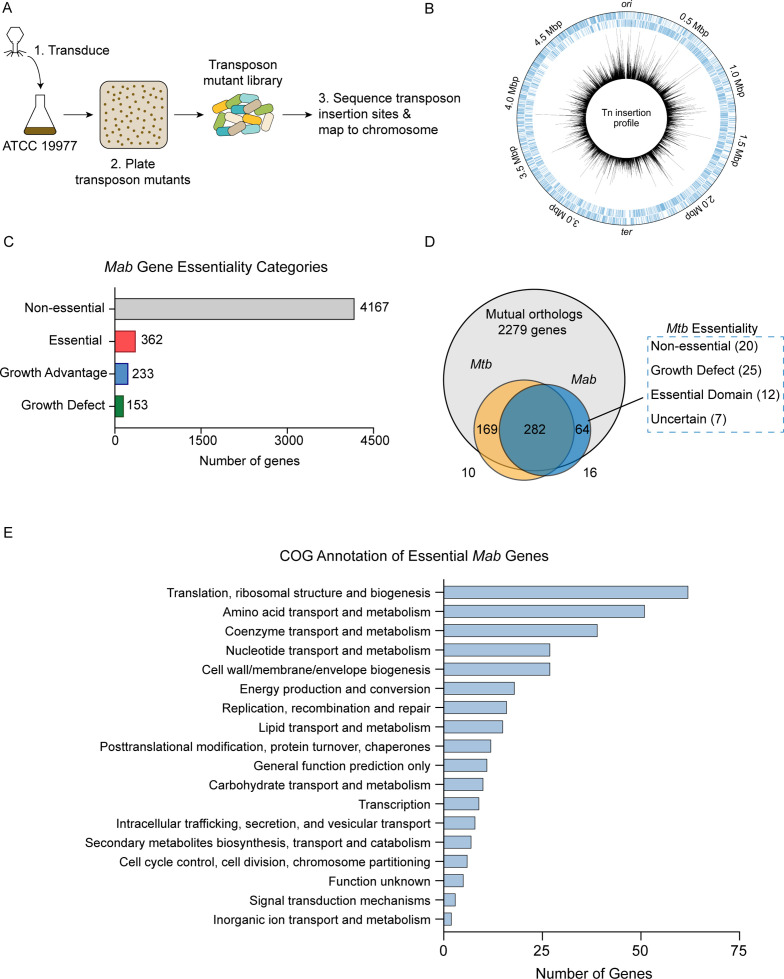
Figure 1. Overview of transposon sequencing (TnSeq) analysis.
(A) Schematic of TnSeq protocol. Phage carrying Himar1 transposon was used to transduce Mycobacterium abscessus (Mab) subsp. abscessus ATCC 19977 cultures. Over 51,000 independent transposon mutants were generated across three libraries. (B) Location of transposon insertions in Mab genome. Black lines represent the average number of transposon insertions per gene across the three replicates. (C) Breakdown of gene essentiality categories as determined by the hidden Markov model (HMM) in TRANSIT. (D) Essentiality comparison of mutual orthologs between Mab and Mycobacterium tuberculosis (Mtb). (E) Clusters of orthologous group (COG) categories of essential genes in Mab.